For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLSMSNAPEDIGDRILQAAASCVRDYGVDRVTLAEIARRAGVSRPTVYRRWPDTRSIISALLTSHVTDVV RAVFLDGHDRQTVVRQVVAVADRLRGDELVKALMHSDLARVYITERLGASQHFLLEGLAARLQVAQGTGS VRAGDPRKLAAMVLLITQSTIQSVDIIDPILDSEALAAELTYSLNGYLA
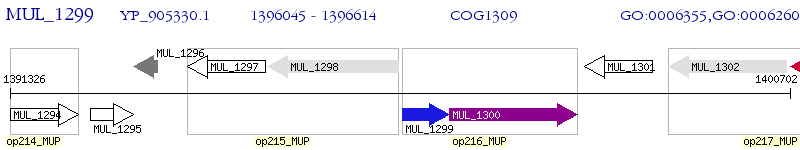
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1299 | - | - | 100% (189) | transcriptional regulatory protein |
| M. ulcerans Agy99 | MUL_4837 | - | 5e-06 | 30.00% (90) | transcriptional regulatory protein |
| M. ulcerans Agy99 | MUL_3128 | - | 1e-05 | 26.88% (160) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2274c | - | 9e-75 | 74.47% (188) | transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2773 | - | 3e-50 | 53.68% (190) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2250c | - | 1e-75 | 75.00% (188) | transcriptional regulator |
| M. leprae Br4923 | MLBr_00717 | - | 7e-05 | 37.50% (56) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3883c | - | 2e-36 | 43.96% (182) | TetR family transcriptional regulator |
| M. marinum M | MMAR_3343 | - | 1e-100 | 99.47% (189) | transcriptional regulatory protein |
| M. avium 104 | MAV_2187 | - | 1e-69 | 69.43% (193) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_4333 | - | 1e-59 | 61.46% (192) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_1508 | - | 3e-50 | 55.00% (180) | Possible transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3760 | - | 5e-52 | 54.74% (190) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2773|M.gilvum_PYR-GCK MMSIRNSE---------------PVEQRILDAAAECIRAYGVERTTVTEI
Mvan_3760|M.vanbaalenii_PYR-1 MVSIRNPER--------------SVEQRILDAAAACILAYGVERTTMTEI
TH_1508|M.thermoresistible__bu ------------------------VEDRILDAAAASVLAVGVNRVTLTDI
MUL_1299|M.ulcerans_Agy99 MLSMSNAP--------------EDIGDRILQAAASCVRDYGVDRVTLAEI
MMAR_3343|M.marinum_M MLSMSNAP--------------EDIGDRILQAAASCVRDYGVDRVTLAEI
Mb2274c|M.bovis_AF2122/97 MLSMSNDR--------------ADTGGRILRAAASCVVDYGVDRVTLAEI
Rv2250c|M.tuberculosis_H37Rv MLSMSNDR--------------ADTGGRILRAAASCVVDYGVDRVTLAEI
MAV_2187|M.avium_104 MLSIRNDRP----------DAALDTGERILAAAASCVVDFGVDRVTLAEI
MSMEG_4333|M.smegmatis_MC2_155 MVSISNDDA----------G--ESLGERILEAAASCVMAFGVDRVTLAEI
MAB_3883c|M.abscessus_ATCC_199 MAPLVDAGGGVTVQAMDAQASADPVTERILDAALSCVTEFGVRRTTLVEV
MLBr_00717|M.leprae_Br4923 MTTARRRLS------------PEDRRAELLALGAEVFGKRPYDEVRIDEI
.:* . . .. : ::
Mflv_2773|M.gilvum_PYR-GCK ARRAGVSRPTIYRRWPDIRWVIAEVLTVRISDVLAAVPAAGPGREELVDR
Mvan_3760|M.vanbaalenii_PYR-1 ARRARVSRPTIYRRWPDIRWVIAELLTVRITGVLEAVPDRGAGRESVVGR
TH_1508|M.thermoresistible__bu ARRARVSRPTIYRRWPDTNALLASLLTTRITAVLDEVPTRGNDRESLVER
MUL_1299|M.ulcerans_Agy99 ARRAGVSRPTVYRRWPDTRSIISALLTSHVTDVVRAVFLDGHDRQTVVRQ
MMAR_3343|M.marinum_M ARRAGVSRPTVYRRWPDTRSIISALLTSHVTDVVRAVSLDGHDRQTVVRQ
Mb2274c|M.bovis_AF2122/97 ARRAGVSRPTVYRRWPDTRSIMASMLTSHIAAVLREVPLDGDDREALVKQ
Rv2250c|M.tuberculosis_H37Rv ARRAGVSRPTVYRRWPDTRSIMASMLTSHIADVLREVPLDGDDREALVKQ
MAV_2187|M.avium_104 ARRAGVSRPTVYRRWPDTRSIVAALLTRHVTDVMRAAPLLGDDRESLVRQ
MSMEG_4333|M.smegmatis_MC2_155 ARRAGVSRPTVYRRFPDTRSILAALLTARIVRVLDEVESDARGREALVRR
MAB_3883c|M.abscessus_ATCC_199 AKRAGVSRPSVYRRWPDVRTLVAELLTREMGSILPATG-RGCARERLVRS
MLBr_00717|M.leprae_Br4923 AERAGVSRALMYHYFPDKRAFFAAVVKDEADRLYAATNIPPPAGLTMFEE
*.** ***. :*: :** . ..: ::. . : . :.
Mflv_2773|M.gilvum_PYR-GCK V-------------------VRIAEHLRRDDIVMSVIFNAPTLAMVYITE
Mvan_3760|M.vanbaalenii_PYR-1 V-------------------VAVADHLRADEIVQSVIRNAPGLAMTYITD
TH_1508|M.thermoresistible__bu I-------------------VGFAERLRADEVVMSVLYGAPEFTMVYITD
MUL_1299|M.ulcerans_Agy99 V-------------------VAVADRLRGDELVKALMHS--DLARVYITE
MMAR_3343|M.marinum_M V-------------------VAVADRLRGDELVKALMHS--DLARVYITE
Mb2274c|M.bovis_AF2122/97 I-------------------VAVADRLRGDDLIMSVMHS--ELARVYITE
Rv2250c|M.tuberculosis_H37Rv I-------------------VAVADRLRGDDLIMSVMHS--ELARVYITE
MAV_2187|M.avium_104 I-------------------VTVADLLRQDRLVMSVLHS--ELAPIYITE
MSMEG_4333|M.smegmatis_MC2_155 I-------------------VAVAERLRHDEVIMAVLHTAPDVAMVYIAE
MAB_3883c|M.abscessus_ATCC_199 V-------------------TGLVTQVREHSIFAAILRSDPELLLTYIVQ
MLBr_00717|M.leprae_Br4923 VRVGVLAYMAYHKQNPEAAWAAYVGLGRSDPVLLGIDDEAKNRQMEHIMS
: . . * . :. .: :* .
Mflv_2773|M.gilvum_PYR-GCK RLGTSQQLLIDTLAAAIVQGQAHGSVRAGDPHQMGAMCLLIAQSTIQSAQ
Mvan_3760|M.vanbaalenii_PYR-1 RLGTSQQILIDALAEAIKAGQDEGSIRAGDPRGMAAMCLLITQSTIQSAQ
TH_1508|M.thermoresistible__bu RLGSSQRIMIEALAREIAEAQETGTVRAGDPHQLAAMCGLIAQSAVQSAR
MUL_1299|M.ulcerans_Agy99 RLGASQHFLLEGLAARLQVAQGTGSVRAGDPRKLAAMVLLITQSTIQSVD
MMAR_3343|M.marinum_M RLGASQHFLLEGLAARLQVAQGTGSVRAGDPRKLAAMVLLITQSTIQSVD
Mb2274c|M.bovis_AF2122/97 RLGTSQQVLIEGLAARLTVAQRSGSVRSGDARRLATMVLLIAQSTIQSAD
Rv2250c|M.tuberculosis_H37Rv RLGTSQQVLIEGLAARLTVAQRSGSVRSGDARRLATMVLLIAQSTIQSAD
MAV_2187|M.avium_104 RLGTSQHMLIDALAARLRVAQRNGSVRPGDPVQMATMVLLIAQSTIQSAQ
MSMEG_4333|M.smegmatis_MC2_155 RLGTSQQILIDAVAGEIKIAQEGGSVRAGDARQLATMCLLITQSAIQSAQ
MAB_3883c|M.abscessus_ATCC_199 RLGTSQRALIGWTAMLIVEGQADGSIRAGIPEQMAAMVLLIGQSVLQSAR
MLBr_00717|M.leprae_Br4923 RIGDVVSVVPESKLHPDIERDLRVIVHGWLAFTFEVCRQRIMDPSTDADR
*:* : : :: . : . * :. ::
Mflv_2773|M.gilvum_PYR-GCK MVE----KFLDRDALNAELAHSLNGYLKP------
Mvan_3760|M.vanbaalenii_PYR-1 MVA----ELLDNEALNEELAHSLNGYLKP------
TH_1508|M.thermoresistible__bu IVE----PFLDSEALAAELRRSLNGYLKP------
MUL_1299|M.ulcerans_Agy99 IID----PILDSEALAAELTYSLNGYLA-------
MMAR_3343|M.marinum_M IID----PILDSEALAAELTYSLNGYLA-------
Mb2274c|M.bovis_AF2122/97 IVD----SILDSAALATELTHALNGYLC-------
Rv2250c|M.tuberculosis_H37Rv IVD----SILDSAALATELTHALNGYLC-------
MAV_2187|M.avium_104 IVE----PLLDAEALATELAYSLNGYLS-------
MSMEG_4333|M.smegmatis_MC2_155 MVA----PILDADALARELAYSLNGYLKP------
MAB_3883c|M.abscessus_ATCC_199 IVA----DILSPDELVDELATAIDGYLKA------
MLBr_00717|M.leprae_Br4923 LADSCAHALLDAIARVPEISKELADAMAIARQPPQ
: :*. *: : . :