For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSSPAEPRQMADGQVSPRRVFMASAVGSAIEFYDFYIYGTAAALVFPAVFFPNLSHSLALFASIATFST AFLARPIGAALFGHYGDRFSRKGTLVVTLITMGLATMAVGLVPGAATIGAAAPTAIVILRLVQGLAVGGE WSGAALLSAEYSPSQNRGRAATAVPIGTSTGLLLSSLVFLIVSLTIGEDSPAFLSWGWRVPFIASGLLVA VGLYIRLQIAETPQFVEAKAHHQLVKSPLRHVITESPRTLVLTSGTMLIIFTLSFMTNTYFPGFARSELH FSRAAILAAGALGAVVLMVSATAGGILSDRLGRRTVILSSMTLTLPWTLLVTPLLTSGPFWAYLAAMALT YVLFGLAFGPMASFLPESYPVGTRYSGTGVAFNVAGVFGGAIPPLIAGPLQNTLGSWAVGLMLLVICAIS VASISGMRETWAGSAR
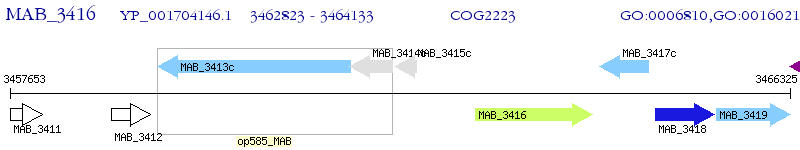
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3416 | - | - | 100% (436) | membrane transporter |
| M. abscessus ATCC 19977 | MAB_4485 | - | 8e-67 | 33.73% (424) | putative transporter |
| M. abscessus ATCC 19977 | MAB_2481 | - | 5e-64 | 36.18% (398) | putative integral membrane transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1232 | - | 1e-113 | 48.69% (421) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_2383 | - | 4e-74 | 36.24% (425) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv1200 | - | 1e-113 | 48.69% (421) | integral membrane transport protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_4238 | - | 1e-107 | 48.68% (417) | integral membrane transport protein |
| M. avium 104 | MAV_1344 | - | 1e-111 | 48.24% (425) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_0716 | - | 4e-77 | 38.46% (416) | major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_3714 | - | 2e-71 | 35.93% (423) | PUTATIVE major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_0949 | - | 1e-106 | 48.44% (417) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_2899 | - | 6e-68 | 34.95% (412) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2383|M.gilvum_PYR-GCK ----MGPGGRT---RVSPRRVAIASFVGTTVEFYDFLIYGTAAALVFPKL
TH_3714|M.thermoresistible__bu ----MSAEAATPTSRVTPRRVAVASFIGTTVEFYDFLIYGTAAALVFPKL
MSMEG_0716|M.smegmatis_MC2_155 -MTTIDNEVGQKVGAVSPARVATASFVGTAIEWYDFFIFGTAAGLVFGVA
Mvan_2899|M.vanbaalenii_PYR-1 ------MNEASVADSALLRRVALSSLLGTAIEYYDFLVYGTMSALVFGLV
Mb1232|M.bovis_AF2122/97 -----------------MKRVALACLVGSAIEFYDFLIYGTAAALVFPTV
Rv1200|M.tuberculosis_H37Rv -----------------MKRVALACLVGSAIEFYDFLIYGTAAALVFPTV
MMAR_4238|M.marinum_M -----------------MSRVALACVVGSTVEYYDFCIYGTAAALVFPAV
MUL_0949|M.ulcerans_Agy99 -----------------MSRVALACVVGSTVEYYDFCIYGTAAALVFPAV
MAV_1344|M.avium_104 ---MGEPSARATTPGTPMRRIAAACLVGSAIEFYDFLIYGTAAALVFPAV
MAB_3416|M.abscessus_ATCC_1997 MTSSPAEPRQMADGQVSPRRVFMASAVGSAIEFYDFYIYGTAAALVFPAV
*: :. :*:::*:*** ::** :.***
Mflv_2383|M.gilvum_PYR-GCK FFPNASPAAGILLSFATFGVGFIARPLGGIVFGHFGDRVGRRRMLVYSLV
TH_3714|M.thermoresistible__bu FFPTASPASGVLLSFATFGVGFFARPLGGIVFGHFGDRLGRKRMLVYSLV
MSMEG_0716|M.smegmatis_MC2_155 FFPNLSPLSGTLAAFATFGVAFFSRPAGAIVFGHFGDRIGRKKLLVLSLL
Mvan_2899|M.vanbaalenii_PYR-1 FFPDSDPAVATIAAFGTLAAGYVARPVGGIVFGHFGDRIGRKSMLVLTMT
Mb1232|M.bovis_AF2122/97 FFPHLDPTVAAVASMGTFAVAFLSRPFGAAVFGYFGDRLGRKKTLVATLL
Rv1200|M.tuberculosis_H37Rv FFPHLDPTVAAVASMGTFAVAFLSRPFGAAVFGYFGDRLGRKKTLVATLL
MMAR_4238|M.marinum_M FYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDRLGRKKTLVATLL
MUL_0949|M.ulcerans_Agy99 FYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDRLGRKKTLVATLL
MAV_1344|M.avium_104 FFPRLGPTVATIASMATFATAFLSRPLGAAVFGYFGDRLGRKKTLVATLL
MAB_3416|M.abscessus_ATCC_1997 FFPNLSHSLALFASIATFSTAFLARPIGAALFGHYGDRFSRKGTLVVTLI
*:* . . . ::.*:...:.:** *. :**::***..*: ** ::
Mflv_2383|M.gilvum_PYR-GCK GMGVSTVLIGVLPTYAQIGLAAPILLTVLRLAQGFAVGGEWGGATLMAVE
TH_3714|M.thermoresistible__bu GMGVATVLIGLLPTYAQIGMAAPALLTLLRLAQGFAMGGEWGGATLMAVE
MSMEG_0716|M.smegmatis_MC2_155 LMGSATVAIGLIPTYDQIGIAAPVLLVVARLVQGFAVGGEWGGAVLMAVE
Mvan_2899|M.vanbaalenii_PYR-1 LMGTASFLIGLLPSFAAVGVAAPILLVALRVIQGVAIGGEWGGATLMVTE
Mb1232|M.bovis_AF2122/97 IMGLATVTVGLVPTTVAIGAAAPLILTTMRLLQGFAVGGEWAGSALLSAE
Rv1200|M.tuberculosis_H37Rv IMGLATVTVGLVPTTVAIGAAAPLILTTMRLLQGFAVGGEWAGSALLSAE
MMAR_4238|M.marinum_M IMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVGGEWAGSALLSIE
MUL_0949|M.ulcerans_Agy99 IMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVGGEWAGSALLSIE
MAV_1344|M.avium_104 IMGASTVSVGLVPSTASIGIAAPLLLTVLRLLQGFAVGAEWAGSVLLSAE
MAB_3416|M.abscessus_ATCC_1997 TMGLATMAVGLVPGAATIGAAAPTAIVILRLVQGLAVGGEWSGAALLSAE
*. ::. :*::* :* :** :. *: **.*:*.**.*:.*: *
Mflv_2383|M.gilvum_PYR-GCK HATADRRGFYGAFPQMGAPAQTAAATLAFYLAARLPDDQ---FLAWGWRI
TH_3714|M.thermoresistible__bu HASADRKGFYGAFPQMGAPAGTATATLAFFAVSRLPDGQ---FLSWGWRL
MSMEG_0716|M.smegmatis_MC2_155 HAPPKRRAFYGSFPQAGVPAGLVLATVAFLIVERLPDDE---IMAWGWRV
Mvan_2899|M.vanbaalenii_PYR-1 HAEAHRRGFWNGVMQMGSPIGSLLSVAVVTVITLLPDES---FLSWGWRI
Mb1232|M.bovis_AF2122/97 YAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGESSPTFMQWGWRI
Rv1200|M.tuberculosis_H37Rv YAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGESSPTFMQWGWRI
MMAR_4238|M.marinum_M SAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSAFLQWGWRV
MUL_0949|M.ulcerans_Agy99 SAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSALLQWGWRV
MAV_1344|M.avium_104 YAPTDRRGWYGMFTLLGGGTAGILASLTFLAVNLTMGEHSPAFMHWGWRV
MAB_3416|M.abscessus_ATCC_1997 YSPSQNRGRAATAVPIGTSTGLLLSSLVFLIVSLTIGEDSPAFLSWGWRV
: . .:. * : .. . :: ****:
Mflv_2383|M.gilvum_PYR-GCK PFLVSAILIVIGLVVRLTVTESPHFAE----LQRRSE--VTRTPIVAAFR
TH_3714|M.thermoresistible__bu PFLLSAVLIGIGLAVRLTLTESPDFAA----VQARSA--VHRMPIAEAFR
MSMEG_0716|M.smegmatis_MC2_155 PFLASSALLLVGLYVRLQITESPEFEQ----IRRNDQ--VVRRPIVDVLR
Mvan_2899|M.vanbaalenii_PYR-1 PFLVSLLLLAVGIYVRLSVTESPVFEA----ARKEAAGNAPRVPFVEILR
Mb1232|M.bovis_AF2122/97 PFLVSAALIAVALYVRFNIDETPVFAR--ERADEKTRLGPAETPIAQVLR
Rv1200|M.tuberculosis_H37Rv PFLVSAALIAVALYVRFNIDETPVFAR--ERADEKTRLGPAETPIAQVLR
MMAR_4238|M.marinum_M PFLLSALLIAFALYVRLEINETPVFARAMAQADDQESTAAARAPLAAVLC
MUL_0949|M.ulcerans_Agy99 PFLLSALLIAFALYVRLEINETPVFARAMAQADDQESTAAARAPLAAVLC
MAV_1344|M.avium_104 PFLISSGLIGIALYVRLNIDETPIFVE------EKARHLVPKAPLTELLR
MAB_3416|M.abscessus_ATCC_1997 PFIASGLLVAVGLYIRLQIAETPQFVE------AKAHHQLVKSPLRHVIT
**: * *: ..: :*: : *:* * . . *: :
Mflv_2383|M.gilvum_PYR-GCK RHWRQILLVAGAYLSQGIFAYICVAYLVSYATTVAEIPRDWALLGVFLGA
TH_3714|M.thermoresistible__bu RHRRQILLVAGAYLSQGVFAYICVAYLVSYGTSVAGIDRTATLFGVSVAA
MSMEG_0716|M.smegmatis_MC2_155 SHKRPLAVGILTQAASLVPFYLVTVFVLSHGPEELGVARETILMSLLVAC
Mvan_2899|M.vanbaalenii_PYR-1 RP-RTLVLACAVGIGPFALTALISTYMISYAT-AIGYHRTDVMTALIFTS
Mb1232|M.bovis_AF2122/97 RQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSILFDSVLGG
Rv1200|M.tuberculosis_H37Rv RQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSILFDSVLGG
MMAR_4238|M.marinum_M RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVLGG
MUL_0949|M.ulcerans_Agy99 RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVLGG
MAV_1344|M.avium_104 LQRREIILVAGSFVGGMGFIYLGNTFLVMYAHNHLGYSRSFIWGIGALGG
MAB_3416|M.abscessus_ATCC_1997 ESPRTLVLTSGTMLIIFTLSFMTNTYFPGFARSELHFSRAAILAAGALGA
* : : : .:. .. * .
Mflv_2383|M.gilvum_PYR-GCK VVAVATYPLFGALSDRVGRKPLYLAGVIAMALAVVPTFALINTGRPVMFV
TH_3714|M.thermoresistible__bu VLAVVMYPLFGSLSDTFGRKPLFLAGVIAMGVSIGPAFALIDTGRPALFL
MSMEG_0716|M.smegmatis_MC2_155 VLDIFTVPYVSIVADRMGPRKMLILGSLYMAAIGYPFFWLLSTGEGWAVL
Mvan_2899|M.vanbaalenii_PYR-1 LTGLIAIPFFSALSDHVGRRLVMVAGAIGIIGYAWPFYALVDM-RSQAYL
Mb1232|M.bovis_AF2122/97 LLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLIDSGSPSLFA
Rv1200|M.tuberculosis_H37Rv LLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLIDSGSPSLFA
MMAR_4238|M.marinum_M LVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFT
MUL_0949|M.ulcerans_Agy99 LVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFT
MAV_1344|M.avium_104 LTSMACVACSAWISDRVGRRRVMLWGLVACLPWAFVVIPLIDTGRPVCYV
MAB_3416|M.abscessus_ATCC_1997 VVLMVSATAGGILSDRLGRRTVILSSMTLTLPWTLLVTPLLTSGPFWAYL
: : . . :.* .* : : : . *:
Mflv_2383|M.gilvum_PYR-GCK LALVLIFGLAMAPAAGVTGALFSMAFDADVRYSGVSVGYTLSQVLGSAFA
TH_3714|M.thermoresistible__bu VALVLVFGLAMAPAAGVTGALFAMTFDADVRYSGVSIGYTLSQVLGSAFA
MSMEG_0716|M.smegmatis_MC2_155 VAMILIVTVGHAVTYSAVAAYVSRLFPADVRYTGASVSYQMGGVVFSAPA
Mvan_2899|M.vanbaalenii_PYR-1 TAAMVIAQLIQSMMYAPLGALFSEMFGTTVRYTGASMGYQLAALLGAGFT
Mb1232|M.bovis_AF2122/97 VAVVGMYAIG-GFGFGPTASFIPELFATSYRYTGSALAANLAGVAGGALP
Rv1200|M.tuberculosis_H37Rv VAVVGMYAIG-GFGFGPTASFIPELFATSYRYTGSALAANLAGVAGGALP
MMAR_4238|M.marinum_M VALLGLYAVA-ASGFGPAASLIPELFATRYRYTGTALALNVAGIVGGAAP
MUL_0949|M.ulcerans_Agy99 VALLGLYAVA-ASGFGPAASLIPELFATRYRYTGTALALNVAGIVGGAAP
MAV_1344|M.avium_104 VAVLGMFGTA-AVANGPTAAFVPELFATRYRYSGAAVAMNLAGIVGAAVP
MAB_3416|M.abscessus_ATCC_1997 AAMALTYVLF-GLAFGPMASFLPESYPVGTRYSGTGVAFNVAGVFGGAIP
* . . .: .. : . **:* .:. :. : .. .
Mflv_2383|M.gilvum_PYR-GCK PTIATALYAATGSSNSIAAYLIAASAVSAVAVCFMPGGWR---AAPAEAR
TH_3714|M.thermoresistible__bu PTIAAALYAATESSGSIVAYLIGVSVISAVSVCLLPGGWGRTGDQPATEV
MSMEG_0716|M.smegmatis_MC2_155 PFIAVTLWSATHGHWSLAAYLAAACLATVFALMFTK-----------TAK
Mvan_2899|M.vanbaalenii_PYR-1 PLIASALHAQEVSRTPLVALAAGCGLVTVVAVWRIS-----------ETR
Mb1232|M.bovis_AF2122/97 PVIAGALVATYGS-WAIGVMLAILALISLVCTYRLP-----------ETA
Rv1200|M.tuberculosis_H37Rv PVIAGALVATYGS-WAIGVMLAILALISLVCTYRLP-----------ETA
MMAR_4238|M.marinum_M PLIAGTLLASYGS-SAVGLMMTALVAASLLCTYLLP-----------ETA
MUL_0949|M.ulcerans_Agy99 PLIAGTLLASYGS-SAVGLMMTALVAASLLCTYLLP-----------ETA
MAV_1344|M.avium_104 PLLAGTLLATYGS-WAIGLMMASLVLASFVSVYLLP-----------ETR
MAB_3416|M.abscessus_ATCC_1997 PLIAGPLQNTLGS-WAVGLMLLVICAISVASISGMR-----------ETW
* :* .* . .: : .
Mflv_2383|M.gilvum_PYR-GCK PEHTPTAPTG------
TH_3714|M.thermoresistible__bu PASVRPMVSGPD----
MSMEG_0716|M.smegmatis_MC2_155 VSY-------------
Mvan_2899|M.vanbaalenii_PYR-1 GTDLTTV---------
Mb1232|M.bovis_AF2122/97 GSALVSR---------
Rv1200|M.tuberculosis_H37Rv GSALVSR---------
MMAR_4238|M.marinum_M GAPLTAA---------
MUL_0949|M.ulcerans_Agy99 GAPLTAA---------
MAV_1344|M.avium_104 GAALDAAAAAEKVAAR
MAB_3416|M.abscessus_ATCC_1997 AGSAR-----------