For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
CGRLLASAPPKWRKPVPEAVIVAALRTPIGTARKGTLRDTNAFDLAHHVVSEATADLDPGQIDDVILGEG LYGGGVIARHAAITAGLGHVPGLAQNRHCAAGQAAVQGAAASVRAGMDQLIIAGGVNSASTSPRPRMQID GEWVDWFPPTHPDRPDAPNMDMSITVGWNAAVAAGVSRAEMDAWALRSHRNAIAAIDEGRFKEEIVPIPT PHGLFCVDEHPRRDTTMEKLAALKPLHPEIDGFSITAGNACGANDGAAVLTIASDRMAQGLGMPALARVL SWASVGVDPAATGLAPVEAIPKALARGHLSISDVDLFEINEAFASMCVATVKLLELDADRVNVSGSGCSL GHPVAATGARMLVTLVHELRRRGGGIGLAAMCAGGGMGSATVIEVAAP*
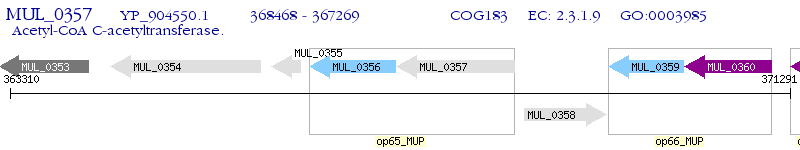
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0357 | - | - | 100% (399) | beta-ketoacyl CoA thiolase |
| M. ulcerans Agy99 | MUL_0207 | fadA3 | 7e-66 | 38.86% (404) | acetyl-CoA acetyltransferase |
| M. ulcerans Agy99 | MUL_2563 | fadA6_4 | 7e-58 | 38.69% (398) | acetyl-CoA acetyltransferase FadA6_4 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1103c | fadA3 | 1e-67 | 38.82% (407) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_4356 | - | 1e-109 | 53.49% (387) | acetyl-CoA acetyltransferase |
| M. tuberculosis H37Rv | Rv1074c | fadA3 | 1e-67 | 38.82% (407) | acetyl-CoA acetyltransferase |
| M. leprae Br4923 | MLBr_02162 | fadA | 2e-58 | 38.24% (408) | acetyl-CoA acetyltransferase |
| M. abscessus ATCC 19977 | MAB_1192c | - | 4e-67 | 38.82% (407) | acetyl-CoA acetyltransferase |
| M. marinum M | MMAR_4788 | - | 0.0 | 100.00% (383) | beta-ketoacyl CoA thiolase |
| M. avium 104 | MAV_2127 | - | 1e-108 | 52.97% (387) | acetyl-CoA C-acyltransferase |
| M. smegmatis MC2 155 | MSMEG_2224 | - | 1e-113 | 55.22% (393) | acetyl-CoA C-acyltransferase |
| M. thermoresistible (build 8) | TH_0166 | fadA6 | 6e-60 | 40.57% (387) | PROBABLE ACETYL-CoA ACETYLTRANSFERASE FADA6 |
| M. vanbaalenii PYR-1 | Mvan_1988 | - | 1e-110 | 53.45% (391) | acetyl-CoA acetyltransferases |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0357|M.ulcerans_Agy99 MCGRLLASAPPKWRKPVPEAVIVAALRTPIGTARKGTLRDTNAFDLAHHV
MMAR_4788|M.marinum_M ----------------MPEAVIVAALRTPIGTARKGTLRDTNAFDLAHHV
Mflv_4356|M.gilvum_PYR-GCK ----------------MSTPVIVGAARTAIGRSFKGTLVNTPPETLITTV
Mvan_1988|M.vanbaalenii_PYR-1 ----------------MPTPVIVGAARTAIGRSFKGTLVNTPPETLITTV
MSMEG_2224|M.smegmatis_MC2_155 -------------MKIMPKPVIVGAARTAIGRSFKGTLVNTPPETLITTV
MAV_2127|M.avium_104 ----------------MGTPVIVGAVRTAIGRSFKGTLVNTPPETLITAV
Mb1103c|M.bovis_AF2122/97 ----------------MPEAVIVSTARSPIGRAMKGSLVGMRPDDLAVQM
Rv1074c|M.tuberculosis_H37Rv ----------------MPEAVIVSTARSPIGRAMKGSLVGMRPDDLAVQM
MAB_1192c|M.abscessus_ATCC_199 ----------------MPEAVIVSTARSPIGRAGKGSLVSMRPDDLAAQM
MLBr_02162|M.leprae_Br4923 ---------------MSEEAFIYEAIRTPRGKQKNGSLHEVKPVSLVVGL
TH_0166|M.thermoresistible__bu --------------MAMTEAYIVDAVRTPVGK-RNGGLSGAHPADMAAHV
. * : *:. * :* * . : :
MUL_0357|M.ulcerans_Agy99 VS-EAT--ADLDPGQIDDVILGEGLYGG---GVIARHAAITAGL-GHVPG
MMAR_4788|M.marinum_M VS-EAT--ADLDPGQIDDVILGEGLYGG---GVIARHAAITAGL-GHVPG
Mflv_4356|M.gilvum_PYR-GCK LP-EVIRRSGVDPADIDDIIFAESHYGG---GDLARYAADAIGL-QHVPG
Mvan_1988|M.vanbaalenii_PYR-1 LP-EVVRRSGVDPADIDDIIFAESHYGG---GDLARYAADAVGL-QHVPG
MSMEG_2224|M.smegmatis_MC2_155 LP-EIVRRSGVDPADIDDIIFAESHYGG---GDLARYAADATGL-QHVPG
MAV_2127|M.avium_104 LP-EVIRRSGVDPAAIDDIIFAESHYGG---GDLARYAAAATGL-EDIPG
Mb1103c|M.bovis_AF2122/97 VRAALDKVPALNPHQIDDLMMGCGLPGGESGFNIARVVAVALGY-DFLPG
Rv1074c|M.tuberculosis_H37Rv VRAALDKVPALNPHQIDDLMMGCGLPGGESGFNIARVVAVALGY-DFLPG
MAB_1192c|M.abscessus_ATCC_199 VRAALDKVPALDPKDVDDLMLGCGQPGGEAGFNIGRAVAVQLGY-DFLPG
MLBr_02162|M.leprae_Br4923 VEELRKRNPDLDENLISDVILGCVSPVGDQGGDIARATVLAAGLPVTAGG
TH_0166|M.thermoresistible__bu IKTLVGRH-DFDPAAIDDVIFGCLDNIGSQAGDIARTAALAAGLPESVPG
: .: :.*:::. * :.* .. * *
MUL_0357|M.ulcerans_Agy99 LAQNRHCAAGQAAVQGAAASVRAGMDQLIIAGGVNSASTSPRPRMQ----
MMAR_4788|M.marinum_M LAQNRHCAAGQAAVQGAAASVRAGMDQLIIAGGVNSASTSPRPRMQ----
Mflv_4356|M.gilvum_PYR-GCK QSVNRHCAGSLTAIGNAAAQIGSGMERVMIAGGVQSLSMTPLVNWR----
Mvan_1988|M.vanbaalenii_PYR-1 QSVNRHCAGSLTAIGNASAQIGSGMERVLIAGGVQSLSMTPLVNWR----
MSMEG_2224|M.smegmatis_MC2_155 QSVNRHCAGSLTAIGNAAAQIGSGMERVLIAGGVQSLSMTPLVNWR----
MAV_2127|M.avium_104 QSVNRHCAGSLTAIGNAAAQIGSGMERVLIAGGVQSLSMTPLVNWR----
Mb1103c|M.bovis_AF2122/97 TTVNRYCSSSLQTTRMAFHAIKAGEGDAFISAGVETVSRFAKGNSDSWPD
Rv1074c|M.tuberculosis_H37Rv TTVNRYCSSSLQTTRMAFHAIKAGEGDAFISAGVETVSRFAKGNSDSWPD
MAB_1192c|M.abscessus_ATCC_199 TTVNRYCSSSLQTTRMAFHAIKAGEGHAFISAGVETVSRFPKGTSDGWPD
MLBr_02162|M.leprae_Br4923 VQLNRFCASGLEAVNTAAQKVRSGWDDLVLAGGVESMSRVPMGSD-----
TH_0166|M.thermoresistible__bu VTIDRQCGSAQQAVHFAAQGVMSGTVDLIVAGGVQKMSQFPILSA-----
:* *... : * : :* .::.**:. * .
MUL_0357|M.ulcerans_Agy99 -------IDG---EWVD-WFPPTHPDRPDAPNMDMSITVGWN---AAVAA
MMAR_4788|M.marinum_M -------IDG---EWVD-WFPPTHPDRPDAPNMDMSITVGWN---AAVAA
Mflv_4356|M.gilvum_PYR-GCK -------IPGPELKFEERWMPPTHVETPDAPTKDMSITVGWN---TAQAA
Mvan_1988|M.vanbaalenii_PYR-1 -------IPGPELKFEERWMPPTHVETPDAPTKDMSITVGWN---TAQAA
MSMEG_2224|M.smegmatis_MC2_155 -------IPGPELKFEERWMPPTHVETPDAPTKDMSITVGWN---TAQAA
MAV_2127|M.avium_104 -------IPGPELKFEERWMPPTHVEAPDAPTRDMSITVGWN---TAKTV
Mb1103c|M.bovis_AF2122/97 TKNPLFDGAQERSAAAAAGADEWHDPRTDQKLPDIYIAMGQTAENVAIMT
Rv1074c|M.tuberculosis_H37Rv TKNPLFDGAQERSAAAAAGADEWHDPRTDQKLPDIYIAMGQTAENVAIMT
MAB_1192c|M.abscessus_ATCC_199 SHNPLYADAEARTATAAQGATEWHDPRNDGLTPDVYIAMGQTAENVALHT
MLBr_02162|M.leprae_Br4923 -----GGAMGLDPATN-----------YDVMF----VPQGIGADLIATIE
TH_0166|M.thermoresistible__bu -----FAAGEPYGATDPWTGCEGWQARYGDQE----ISQFRGAEMIAEHW
. :. *
MUL_0357|M.ulcerans_Agy99 GVSRAEMDAWALRSHRNAIAAIDEGRFKEEIVPIPTPHG-----LFCVDE
MMAR_4788|M.marinum_M GVSRAEMDAWALRSHRNAIAAIDEGRFKEEIVPIPTPHG-----LFCVDE
Mflv_4356|M.gilvum_PYR-GCK GITREDMDAWAARSHQRAVAAQDDGKFLDEIVPLKVEQFDGSVIDFSVDE
Mvan_1988|M.vanbaalenii_PYR-1 GITREEMDVWAVRSHQRAVAAIDAGKFVDEIVPLKVTQFDGSVIDFSVDE
MSMEG_2224|M.smegmatis_MC2_155 GISREEMDAWAARSHQRAIAAIDAGKFLDEIVPLKVTQFDGSVIDFSVDE
MAV_2127|M.avium_104 GITREEMDAWAARSHQRAIAAIDAGKFVDEILPLKITQADGSVTEFAVDE
Mb1103c|M.bovis_AF2122/97 GISREEQDRWGVRSQNRAEEAIKNGFFEREITPVTLPDG----TTVSTDD
Rv1074c|M.tuberculosis_H37Rv GISREEQDRWGVRSQNRAEEAIKNGFFEREITPVTLPDG----TTVSTDD
MAB_1192c|M.abscessus_ATCC_199 GISREDQDHWGVRSQNRAEKAIADGFFEREITPVTLADG----TVVAKDD
MLBr_02162|M.leprae_Br4923 GFSRDDVDVYALRSQEKAAAAWSGGYFAKSVVPVRDQNGL---LILDHDE
TH_0166|M.thermoresistible__bu KLSREDNEKFALESHRRAVAAIEAGYFDKEITPYADAT---------VDE
.:* : : :. .*:..* * * * .: * *:
MUL_0357|M.ulcerans_Agy99 HPRRDTTMEKLAALKPLHPEIDG-------------------FSITAGNA
MMAR_4788|M.marinum_M HPRRDTTMEKLAALKPLHPEIDG-------------------FSITAGNA
Mflv_4356|M.gilvum_PYR-GCK QPRRDTSVEKLAGLKVLHPEIEG-------------------FSITAGNA
Mvan_1988|M.vanbaalenii_PYR-1 SPRRDTTLEKLAGLKVLHPEIEG-------------------FSITAGNA
MSMEG_2224|M.smegmatis_MC2_155 HPRRDTTVEKLAGLKVLHPEIEG-------------------FSITAGNS
MAV_2127|M.avium_104 HPRRDTTVEKLAGLKVLHPEIEG-------------------FSITAGNS
Mb1103c|M.bovis_AF2122/97 GPRPGTTYEKVSELKPAFRP---------------------NGTVTAGNA
Rv1074c|M.tuberculosis_H37Rv GPRPGTTYEKVSELKPAFRP---------------------NGTVTAGNA
MAB_1192c|M.abscessus_ATCC_199 GPRAGTTYEAISQLKPVFRP---------------------NGTITAGNA
MLBr_02162|M.leprae_Br4923 HMRPDTSKEGLAKLKPAFEGLAAMGGFDDVALQKYHWVEKINHVHTGGNS
TH_0166|M.thermoresistible__bu GPRPDTSLEKMASLPTLIEG----G------------------VLTAAVA
* .*: * :: * *.. :
MUL_0357|M.ulcerans_Agy99 CGANDGAAVLTIASDRMAQGLGMPALARVLSWASVGVDPAATGLAPVEAI
MMAR_4788|M.marinum_M CGANDGAAVLTIASDRMAQGLGMPALARVLSWASVGVDPAATGLAPVEAI
Mflv_4356|M.gilvum_PYR-GCK SGTNDAAAGVALVEADYAAANGLTVMAKVRSWGAVGVAPRDTGLGGVKVI
Mvan_1988|M.vanbaalenii_PYR-1 SGTNDAAAGVALVDADYAAANGLNVMAKVKAWGAVGVAPRDTGLGGVKVI
MSMEG_2224|M.smegmatis_MC2_155 SGTNDAAAGVAVVDSEYAAANGLNVMATVRAWGAVGVPPRDTGLGGVRVI
MAV_2127|M.avium_104 SGTNDAAAAVALVDREYAEAEKLNVMGTVRAWAAAGVPPRDCGLGAVKVI
Mb1103c|M.bovis_AF2122/97 CPLNDGAAAVVITSDTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAS
Rv1074c|M.tuberculosis_H37Rv CPLNDGAAAVVITSDTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAS
MAB_1192c|M.abscessus_ATCC_199 CPLNDGAAALVVMSDVRAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAV
MLBr_02162|M.leprae_Br4923 SGIVDGAALVMIGSEAVGKSQGLTPRARMVATATSGADPVIMLTGPTPAT
TH_0166|M.thermoresistible__bu SQISDGAAALLVASKDAVDRFNLTPRARIHHMSVRGADPVMMLTAPIPAT
. *.** : : . : . : . * * . .
MUL_0357|M.ulcerans_Agy99 PKALARGHLSISDVDLFEINEAFASMCVATVKLLELDADRVNVSGSGCSL
MMAR_4788|M.marinum_M PKALARGHLSISDVDLFEINEAFASMCVATVKLLELDADRVNVSGSGCSL
Mflv_4356|M.gilvum_PYR-GCK GRVLDKAGLKPSDVTLWEINEAFASVPIAAVREYGIDEELVNFSGSGCSL
Mvan_1988|M.vanbaalenii_PYR-1 GRVLDRAGLKPSDVALWEINEAFASVPIAACKEYGIDEELVNFSGSGCSL
MSMEG_2224|M.smegmatis_MC2_155 GKVLDRAGLKPSDVALWEINEAFASVPIAACKEYGLDEELVNFNGSGCSL
MAV_2127|M.avium_104 GKVLQRAGLSPDDVALWEINEAFASVPIAACREYGLDEEKVNFSGSGCSL
Mb1103c|M.bovis_AF2122/97 KKALERAGMAITDIDLVEINEAFAVQVLGSARELGIDEDKLNISGGAIAL
Rv1074c|M.tuberculosis_H37Rv KKALERAGMAITDIDLVEINEAFAVQVLGSARELGIDEDKLNISGGAIAL
MAB_1192c|M.abscessus_ATCC_199 KRALANAGKSIGDIDLFEINEAFAVQVLGSARALGIDEDKLNVSGGAIAL
MLBr_02162|M.leprae_Br4923 RKVLDRAGLTVDDIDLFELNEAFASVVLKFQKDLNIPDEKLNVNGGAIAM
TH_0166|M.thermoresistible__bu QHALKRAGMSIDDIDAVEINEAFAPVVLAWLAELGADPARVNPNGGAIAL
:.* .. *: *:***** : :* .*.. ::
MUL_0357|M.ulcerans_Agy99 GHPVAATGARMLVTLVHELRRRGGGIGLAAMCAGGGMGSATVIEVAAP
MMAR_4788|M.marinum_M GHPVAATGARMLVTLVHELRRRGGGIGLAAMCAGGGMGSATVIEVAAP
Mflv_4356|M.gilvum_PYR-GCK GHPIAASGARMITTLIYELQRRGGGIGVAAMCAGGGQGGGVVIEV---
Mvan_1988|M.vanbaalenii_PYR-1 GHPIAASGARMITTLIYELQRRGGGIGVAAMCAGGGQGGGVVIEV---
MSMEG_2224|M.smegmatis_MC2_155 GHPIAASGARMVTTLVYELQRRGGGIGVAAMCAGGGQGGAVVIEV---
MAV_2127|M.avium_104 GHPIAASGARMVTTLTYELARRGGGIGVAAMCAGGGQGGAVVIEV---
Mb1103c|M.bovis_AF2122/97 GHPFGMTGARITTTLLNNLQTYDKTFGLETMCVGGGQGMAMVIERLA-
Rv1074c|M.tuberculosis_H37Rv GHPFGMTGARITTTLLNNLQTYDKTFGLETMCVGGGQGMAMVIERLA-
MAB_1192c|M.abscessus_ATCC_199 GHPFGMTGARITATLLNNLTTYDKTFGVETMCVGGGQGMAMVIERLA-
MLBr_02162|M.leprae_Br4923 GHPLGATGAMILGTMVDELERCNARRALVTLCIGAGMGVATIIERV--
TH_0166|M.thermoresistible__bu GHPIGCTGARLMTSLLHHLERTGGRYGLQTMCEGGGQANVTIIERL--
***.. :** : :: .* . .: ::* *.* . :**