For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ENEARAGLRSDMRTDSDTDVEPGRDSEARPVRSTLVDQVYERLMELLLNGTLKAGDPVSIDGTSRYLGVS QTPVREALARLESTGLVVRVAMRGYRVPEMPDAKEIADIMDARLLIEPQLAELACNHADPEWLDALAQAV DEQERAPHTADAAAIRRYHRADEQFHRLIAERAGNAALLRVYDALGGHGQRFRLFIGVGVQDSEFAIAEH RELLEAFRRGYGPEVYRIMHAHISSVKRRALSEQARAEHAWTARVQT*
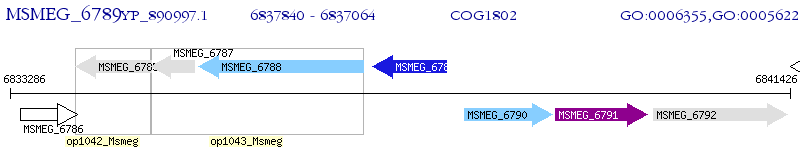
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6789 | - | - | 100% (258) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_2546 | - | 2e-34 | 39.56% (225) | GntR family transcriptional regulator protein |
| M. smegmatis MC2 155 | MSMEG_0535 | - | 2e-14 | 30.14% (209) | GntR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0170c | - | 2e-10 | 31.05% (190) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0958 | - | 4e-15 | 30.05% (213) | GntR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0165c | - | 6e-10 | 29.47% (207) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0068 | - | 5e-15 | 32.82% (195) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0408 | - | 3e-11 | 29.95% (197) | GntR family transcriptional regulator |
| M. avium 104 | MAV_5020 | - | 5e-13 | 30.73% (192) | transcriptional regulator, GntR family protein |
| M. thermoresistible (build 8) | TH_3342 | - | 2e-12 | 33.33% (150) | PUTATIVE transcriptional regulator, GntR family |
| M. ulcerans Agy99 | MUL_1058 | - | 1e-11 | 29.95% (197) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0130 | - | 2e-13 | 33.66% (202) | GntR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0170c|M.bovis_AF2122/97 MIKHDVVWVTLWPERPNNKPPPSPRQVPGNPGPTLKVLASHVNAPLSAKP
Rv0165c|M.tuberculosis_H37Rv MIKHDVVWVTLWPERPNNKPPPSPRQVPGNPGPTLKVLASHVNAPLSAKP
MMAR_0408|M.marinum_M -----------------------------------------MNIPLSARR
MUL_1058|M.ulcerans_Agy99 -----------------------------------------MNILLSARR
MAV_5020|M.avium_104 ---------------------------------------------MSMQP
Mvan_0130|M.vanbaalenii_PYR-1 -----------------------------------------MNAP--ART
TH_3342|M.thermoresistible__bu -------------------------------------------------V
MAB_0068|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6789|M.smegmatis_MC2_155 -------------------------MENEARAGLRSDMRTDSDTDVEPGR
Mflv_0958|M.gilvum_PYR-GCK -------------------------------------------MADAQLQ
Mb0170c|M.bovis_AF2122/97 RSQLPLRRAQLSDEVAGHLRAAIMSGALRSGTFIRLDETAAELGVSVTPV
Rv0165c|M.tuberculosis_H37Rv RSQLPLRRAQLSDEVAGHLRAAIMSGALRSGTFIRLDETAAELGVSVTPV
MMAR_0408|M.marinum_M KSRRPLRRAQLSDEVAGHLRAAIMSGTLRPGTFIRLDETAAELGVSITPV
MUL_1058|M.ulcerans_Agy99 ESRRPLRRAQLSDEVAGHLRAAIMSGTLRPGTFIRLDETAAELGVSITPV
MAV_5020|M.avium_104 RSRRPLRRAQLSDEVAGHLRAAIMSGALRPGTFIRLDETAAELGVSVTPV
Mvan_0130|M.vanbaalenii_PYR-1 PVAGAVRREQLSDEVAARLRAEIMTGALRPDSFIRLDETAARLGVSITPV
TH_3342|M.thermoresistible__bu AVPEFAARPQLAEDVARYVRRRIFDGTYAAGEYVRLDQLAVELGVSVTPV
MAB_0068|M.abscessus_ATCC_1997 --MIRTVAISASEEAYRSVKESILNGDIPGGELLSEGEISARMGCSRTPV
MSMEG_6789|M.smegmatis_MC2_155 DSEARPVRSTLVDQVYERLMELLLNGTLKAGDPVSIDGTSRYLGVSQTPV
Mflv_0958|M.gilvum_PYR-GCK LAALPAAEGRRAEQVMAAVRAALDAGVMRPGVKYSVYQLADALQVSRTPV
::. : : * . : : * ***
Mb0170c|M.bovis_AF2122/97 REALLKLRGEGMVGLEPHRGHVVLPLTRQ-DIDDIFWLQATIAQELATSA
Rv0165c|M.tuberculosis_H37Rv REALLKLRGEGMVGLEPHRGHVVLPLTRQ-DIDDIFWLQATIAQELATSA
MMAR_0408|M.marinum_M REALLKLRGEGMVQLEPHRGHVVLPLTRQ-DIDDIFWLQATIARELAVTA
MUL_1058|M.ulcerans_Agy99 REALLKLRGEGMVQLEPHRGHVVLPLTRQ-DIDDIFWLQATIARELAVTA
MAV_5020|M.avium_104 REALLKLRGEGMVQLEPHRGHVVLPLTRQ-DIEDIFWLQATIAKELAAAA
Mvan_0130|M.vanbaalenii_PYR-1 REALRTLRGEGMVELEPHRGHRVVPLTRT-DIEDIFWLQSTIAQELAATA
TH_3342|M.thermoresistible__bu REALFQLRAEGLLVQQPRRGFMVLPVTER-DLTDIANVQAHVGGELAARA
MAB_0068|M.abscessus_ATCC_1997 REAFLRLETEGWLRLYPKRGALVVPITPE-ERRHVVDARRVIEGAAAARI
MSMEG_6789|M.smegmatis_MC2_155 REALARLESTGLVVRVAMRGYRVPEMPDAKEIADIMDARLLIEPQLAELA
Mflv_0958|M.gilvum_PYR-GCK RDALLRLEEIGLIRFEARQGFRILLPDPR-EIADIVAIRFALEPPAAGRA
*:*: *. * : . :* : : .: : : *
Mb0170c|M.bovis_AF2122/97 TAH-ITDVEIDELDRINNALAGAIGSGDAKTIA---SIEFAFHRVFNKAS
Rv0165c|M.tuberculosis_H37Rv TAH-ITDVEIDELDRINNALAGAIGSGDAKTIA---SIEFAFHRVFNKAS
MMAR_0408|M.marinum_M TER-ISEIDIGELEHINDALTAAVGAGDTEAIA---ALEFAFHRVLNQSS
MUL_1058|M.ulcerans_Agy99 TER-ISEVVIGELEHINDALTAAVGAGDTEAIA---ALEFAFHRVLNQSS
MAV_5020|M.avium_104 TDH-ITDTEIDELDRINDALAAAVGSGDAETIA---GLEFGFHRVFNQAS
Mvan_0130|M.vanbaalenii_PYR-1 ARN-ITDAQIDELAQLNERLAEAVEQRDPDRVA---RAEFTFHRAFNRAG
TH_3342|M.thermoresistible__bu AQN-ITDEQLRVLTAIQDELEEAYTRDDEDRAV---RLNHEFHRAINVAA
MAB_0068|M.abscessus_ATCC_1997 AERGAPQSLLSDLSALIEVQLGRAAQGDAAGFA---AADVDFHRAIVAKL
MSMEG_6789|M.smegmatis_MC2_155 CNH-ADPEWLDALAQAVDEQERAPHTADAAAIRRYHRADEQFHRLIAERA
Mflv_0958|M.gilvum_PYR-GCK AAV-CGPPLADRLAERMELLRRAAARGDEPGFA---AHDLLLHDHILEAA
* : * : :* :
Mb0170c|M.bovis_AF2122/97 RRIKLAWF---LLNAARYMPAQVFAADPRWGADAVNSHRQLIAALRRRDT
Rv0165c|M.tuberculosis_H37Rv RRIKLAWF---LLNAARYMGAGVRGRP----AMGRGRGEQSSAADRR---
MMAR_0408|M.marinum_M GRIKLAWF---LLNAARYMPAQLYASDPQWGAAAVDNHRQLIAALRRRDT
MUL_1058|M.ulcerans_Agy99 GRIKLAWF---LLNAARYMPAQLYASDPQWGAAAVDNHRQLIAALRRRDT
MAV_5020|M.avium_104 GRIKLAWF---LLNAARYMPVLVYAADPQWGRAAVDNHRQLIAALRRRDT
Mvan_0130|M.vanbaalenii_PYR-1 GRIKLAWF---LLHAARYLPPQLFASDETWGPVAVASHAELIEALRRRDV
TH_3342|M.thermoresistible__bu DSPKLAQV---MSQITRYAPESVFPTISGWPARSTEHHRRVLAALADRDP
MAB_0068|M.abscessus_ATCC_1997 GNPLLENFYDSLRERQRRMAAAAIGVDPVRVARSVAGHRALVDALAAGDA
MSMEG_6789|M.smegmatis_MC2_155 GNAALLRVYD-ALGGHGQRFRLFIGVGVQDSEFAIAEHRELLEAFRRGYG
Mflv_0958|M.gilvum_PYR-GCK GNNRARRIVR-SLRESTRLLGGITADRSRTMSDIDAEHQPVVDAVIANDP
. *
Mb0170c|M.bovis_AF2122/97 AAVIEHTVWQFTDGARRLTEALAETEVFG------------------
Rv0165c|M.tuberculosis_H37Rv AAPPRHSRR---NRAHRLAVHRWGTQADGGPG---------------
MMAR_0408|M.marinum_M AAVIEHSVWQFTDAAHRLTQMLETNGIFN------------------
MUL_1058|M.ulcerans_Agy99 AAVIEHSVWQFTDAAHRLTQMLETNGIFN------------------
MAV_5020|M.avium_104 AAVIEHTVWQFTDAAARLTAMRDRTGTPSNRG---------------
Mvan_0130|M.vanbaalenii_PYR-1 DTVVELTRGQFTDGRARLIAWLEQVGLWS------------------
TH_3342|M.thermoresistible__bu DGARAAMSEHLAAGAGPLIEHLKEHGVIAQ-----------------
MAB_0068|M.abscessus_ATCC_1997 ARFSTELVEHMKVVTEVD-----------------------------
MSMEG_6789|M.smegmatis_MC2_155 PEVYRIMHAHISSVKRRALSEQARAEHAWTARVQT------------
Mflv_0958|M.gilvum_PYR-GCK AAAAAAMRTHLTSTGRLLVAQAVRDQDSPLDAAAVWAEAVGANTELP