For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SIKHVSSPPSQHTAAPVPSAVRIGFSRVLPELKMFYRRPEQMVLTFSMPAVICLLLGSIFSDPLPVGGVS TGAVIAASMLAYGILSTSFINLGISIAADRDTGALRRLRGTPTTATSYFIGKIMLVAIVSFAEAVLLMAV GVLVFGLRLPADLFGWFTLAWVFVLGVVSCSLLGVFISNFASNAVAAAALTNGPAVGLQFLSGTYVPLMV LPTWMLAIGSLFPVKWMAQGFRSVLLPPELVAIEPAGSWEHWRIFLVLMAWCVGGVIACRAVFRWSDRD*
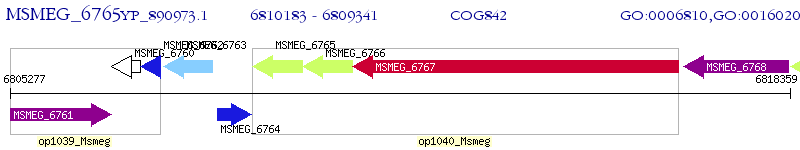
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6765 | - | - | 100% (280) | ABC-2 type transporter superfamily protein |
| M. smegmatis MC2 155 | MSMEG_6510 | - | 2e-14 | 25.71% (280) | ABC-type drug export system, membrane protein |
| M. smegmatis MC2 155 | MSMEG_3118 | - | 5e-13 | 31.15% (183) | ABC transporter efflux protein, DrrB family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1492c | - | 4e-14 | 28.94% (273) | unidentified antibiotic-transport integral membrane ABC |
| M. gilvum PYR-GCK | Mflv_3681 | - | 4e-14 | 29.26% (270) | multidrug ABC transporter inner membrane protein |
| M. tuberculosis H37Rv | Rv1457c | - | 4e-14 | 28.94% (273) | unidentified antibiotic-transport integral membrane ABC |
| M. leprae Br4923 | MLBr_00589 | - | 2e-14 | 27.34% (278) | hypothetical protein MLBr_00589 |
| M. abscessus ATCC 19977 | MAB_2753 | - | 8e-11 | 33.09% (136) | ABC transporter permease |
| M. marinum M | MMAR_2262 | - | 8e-14 | 30.77% (273) | antibiotic ABC transporter transmembrane protein |
| M. avium 104 | MAV_5182 | - | 2e-14 | 25.48% (263) | ABC-type drug export system, membrane protein |
| M. thermoresistible (build 8) | TH_0342 | - | 4e-13 | 28.09% (267) | PROBABLE UNIDENTIFIED ANTIBIOTIC-TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_1858 | - | 5e-14 | 29.89% (271) | antibiotic ABC transporter transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_2729 | - | 4e-12 | 28.14% (263) | multidrug ABC transporter inner membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3681|M.gilvum_PYR-GCK --MSVENR------FAPGTFTPDPR----PAKVPKMLAAQFALELRLLLR
Mvan_2729|M.vanbaalenii_PYR-1 --MTTENR------FTPGTFTPDPR----PAPVPRMLAAQFRLELRLLLR
MAB_2753|M.abscessus_ATCC_1997 MTDTTTPR------FDPGTFTPDPR----PSAVHTMLFAQFWLELKLLLR
TH_0342|M.thermoresistible__bu ----VTNR------FPAGTFTPDPR----PAPVPRMLAAQYGLELKLLLR
Mb1492c|M.bovis_AF2122/97 --MTQTN----RPAFPAGTFSPDPR----PNAVPLMLAAQFSLELKLLLR
Rv1457c|M.tuberculosis_H37Rv --MTQTN----RPAFPAGTFSPDPR----PNAVPLMLAAQFSLELKLLLR
MMAR_2262|M.marinum_M --MTQTNG---EGTFPVGTFTPDPR----PSAVPRMLAAQFGLELKLLLR
MUL_1858|M.ulcerans_Agy99 --MTQTNG---EGTFPVGTFTPDPR----PSAVPRMLAAQFGLELKLLLR
MLBr_00589|M.leprae_Br4923 --MTQNNTLRNSPLFPAGTFTPNPR----PNAVTRMVAAQFMLELKLLLR
MSMEG_6765|M.smegmatis_MC2_155 --MSIKHVS-----SPPSQHTAAPV----PSAVR-IGFSRVLPELKMFYR
MAV_5182|M.avium_104 --MSVDNA-------AAATLVRAPRGWQRVGALSSRVGAFAIVEIQKLQH
. * : : *:: : :
Mflv_3681|M.gilvum_PYR-GCK NGEQLLLTMFIP--ITLLIG---LILLPLGSFGPNRAATFVPAIMALAVI
Mvan_2729|M.vanbaalenii_PYR-1 NGEQLLLTMFIP--ITLLVG---LTLLPLGSFGDNRAATFVPAIMALAVI
MAB_2753|M.abscessus_ATCC_1997 NGEQLLLTMFIP--ITLLIG---LTLLPLGQFPGSRTDIFVPAIMALAVI
TH_0342|M.thermoresistible__bu NGEQLLLTLFIP--ITLLVG---LILLPLGDFGDDRASRFVPAIMALAVI
Mb1492c|M.bovis_AF2122/97 NGEQLLLTMFIP--ITLLVG---LTLLPMGSFGHNRAATFVPVIMALAVI
Rv1457c|M.tuberculosis_H37Rv NGEQLLLTMFIP--ITLLVG---LTLLPMGSFGHNRAATFVPVIMALAVI
MMAR_2262|M.marinum_M NGEQLLLTMFIP--ITLLIG---LTLLPLGSFGQHRAATFVPVIMALAVI
MUL_1858|M.ulcerans_Agy99 NGEQLLLTMFIP--ITLLIG---LTLLPLGSFGQHRAATFVPVIMALAVI
MLBr_00589|M.leprae_Br4923 NGEQLLLTMFIP--ITLLIG---LTLLPLGSFGHHRATTFVPVIMALAVV
MSMEG_6765|M.smegmatis_MC2_155 RPEQMVLTFSMPAVICLLLGS--IFSDPLPVGGVSTGAVIAASMLAYGIL
MAV_5182|M.avium_104 DRTELVTRMVQPALWLLIFGTTFSHLHMIDTGPVSYLAFLAPGIIAQSAL
::: : * *:.* : :.. ::* . :
Mflv_3681|M.gilvum_PYR-GCK STAFTGQAIAVAFDRRYGALKRLGATALPVWGVIAGKSLAVVTVVSLQSV
Mvan_2729|M.vanbaalenii_PYR-1 STAFTGQAIAVAFDRRYGALKRLGATALPVWGIIAGKSLAVGAVVILQSI
MAB_2753|M.abscessus_ATCC_1997 STAFTGQAIAVGFDRRYGALKRLGATALPVWGIIAGKALAVVTVVVLQAA
TH_0342|M.thermoresistible__bu STAFTGQAIAVAFDRRYGALKRLGATALPVWGIIVGKSLAVVTVVFVQSV
Mb1492c|M.bovis_AF2122/97 STAFTGQAIAVAFDRRYGALKRLGATPLPVWGIIAGKSLAVVAVVFLQAI
Rv1457c|M.tuberculosis_H37Rv STAFTGQAIAVAFDRRYGALKRLGATPLPVWGIIAGKSLAVVAVVFLQAI
MMAR_2262|M.marinum_M STAFTGQAIAVAFDRRYGALKRLGATPLPVWGIIAGKSLAVVAVVFLQAI
MUL_1858|M.ulcerans_Agy99 STAFTGQAIAVAFDRRYGALKRLGATPLPVWGIIAGKSLAVVAVVFLQAI
MLBr_00589|M.leprae_Br4923 SSAFTGQAIAVAFDRRYGALKRFGATPLPVWGIIAGKSLAVVTVVFLQAI
MSMEG_6765|M.smegmatis_MC2_155 STSFINLGISIAADRDTGALRRLRGTPTTATSYFIGKIMLVAIVSFAEAV
MAV_5182|M.avium_104 FISIF-YGIQIIWDRDAGVLAKLMVTPAPASALVTGKAFAAGVRSVAQVV
:: .* : ** *.* :: *. .. . . ** : . :
Mflv_3681|M.gilvum_PYR-GCK LFGAIG---FALGWRPPLAGLALGALIIALGTAVFAALGLLLGGTLRAEI
Mvan_2729|M.vanbaalenii_PYR-1 LFGAIG---FALGWRPPLAGLALGAVIIALGTAVFAALGLLLGGTLRAEI
MAB_2753|M.abscessus_ATCC_1997 VIGAIG---GALGWRPHPVGLLVGAVVIAIGTAAFVAMGLLLGGTLRAEI
TH_0342|M.thermoresistible__bu VLGAIG---AALGWRPQPAGLLLGAVIIALGTAVFAALGLLLGGTLRAEI
Mb1492c|M.bovis_AF2122/97 ILGAIG---FALGWRPALTALTLGAGIIALGTAGFAALGLLLGGTLRAEI
Rv1457c|M.tuberculosis_H37Rv ILGAIG---FALGWRPALTALTLGAGIIALGTAGFAALGLLLGGTLRAEI
MMAR_2262|M.marinum_M VLGAIG---FALGWRPALAALALGGAVIALGTAGFAALGLLLGGTLRAEI
MUL_1858|M.ulcerans_Agy99 VLGAIG---FALGWRPALAALALGGAVIALGTAGFAALGLLLGGTLRAEI
MLBr_00589|M.leprae_Br4923 ILGAIG---VMLGWRPALICLALGAAIIALGTAGLAALGLLLGGTLRAEI
MSMEG_6765|M.smegmatis_MC2_155 LLMAVGVLVFGLRLPADLFGWFTLAWVFVLGVVSCSLLGVFISNFASNAV
MAV_5182|M.avium_104 GVLALAY-LMRIGLTVNPLRILAAMGIVMLGAAFFACLSMTLAGLVRNRD
. *:. : :. :*.. :.: :..
Mflv_3681|M.gilvum_PYR-GCK VLALAN------LLWFVFAGLGALVLERG----MVPAAVQWVARLTPSGA
Mvan_2729|M.vanbaalenii_PYR-1 VLALAN------LLWFVFAGLGALTLEAG----VVPSTVQWVARLTPSGA
MAB_2753|M.abscessus_ATCC_1997 VLALAN------LLWFIFAGLGALTLEDGRTTDAIPRAVVLLARLSPSGA
TH_0342|M.thermoresistible__bu VLAVAN------LLWFVFAAFGALTVEPEG--DVVPAAAALVARLTPSGA
Mb1492c|M.bovis_AF2122/97 VLAVAN------LMWFVFAGFGALTLESN----VIPTAFKWVARVTPSGA
Rv1457c|M.tuberculosis_H37Rv VLAVAN------LMWFVFAGFGALTLESN----VIPTAFKWVARVTPSGA
MMAR_2262|M.marinum_M VLAVAN------LMWFVFAGFGALTVETD----VIPTAVKWAARLTPSGA
MUL_1858|M.ulcerans_Agy99 VLAIAN------LMWFVFAGFGALTVETD----VIPTAVKWAARLTPSGA
MLBr_00589|M.leprae_Br4923 VLAVAN------LMWFVFAGLGALTVKMN----MISAVVKWAARLTPSGA
MSMEG_6765|M.smegmatis_MC2_155 AAAALTNGPAVGLQFLSGTYVPLMVLPTWMLAIGSLFPVKWMAQGFRSVL
MAV_5182|M.avium_104 RLMGIGQAITMPLFFASNALYPVELMPTW---------LHVLSKINPLSY
* : : : ::
Mflv_3681|M.gilvum_PYR-GCK LTEALVQAMTLS--VDWFGLLVLAAWGAVAALCALRWFRFV---
Mvan_2729|M.vanbaalenii_PYR-1 LTEALTQAMTSS--MDWFGLAVLAVWGAVAGLCALRWFRFT---
MAB_2753|M.abscessus_ATCC_1997 LSEALAAAMTLS--VDWFAIAVLAVWGALAGYGALRWFKFT---
TH_0342|M.thermoresistible__bu LTDALSQAMTLS--VNWFAVAVLAVWGVLGAVCALRWFRFT---
Mb1492c|M.bovis_AF2122/97 LTEALSQAMTVS--VDWFGIVVLAVWGALAALAALRWFRFT---
Rv1457c|M.tuberculosis_H37Rv LTEALSQAMTVS--VDWFGIVVLAVWGALAALAALRWFRFT---
MMAR_2262|M.marinum_M LTEALSRAMVLS--VDWFGVVVLAVWGTLAALAARRWFRFT---
MUL_1858|M.ulcerans_Agy99 LTEALSRAMVLS--VDWFGVVVLAVWGTLAALAARRWFRFT---
MLBr_00589|M.leprae_Br4923 LTEALSQAMTLS--VDWFGVIILTAWGTLAALAALHWFRFT---
MSMEG_6765|M.smegmatis_MC2_155 LPPELVAIEPAGSWEHWRIFLVLMAWCVGGVIACRAVFRWSDRD
MAV_5182|M.avium_104 EVNALRALLIGTP-FNPVDIAVLAAAAVTGILTASTLLRRLVA-
* . . :* . . . . ::