For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TITPLDAHRQANATSPDLAAVLETIAERGVEFVYFQAVTITGRVVGKVAPAKHFERLAVRGVQQHQTAVA NLQGTREGVLLAGGVNAPEYTAIPDLETFAVLPWDTSFARVFCRLYEPDHLAERAGAEFACDSRGLLRRL HAGFTDRTGLELRTGCEPEMTWQGEGLEAQFRPGSSPAYHIEHLERNRPIVKKVVEYAQALGLDMIEGDY EDEFQVELNFMYDRADLTADRLTTYRQICKQVARELGIQASFMPKPATGMMGNGCHHNFSLWRDGVNVLA EPGVTELHLSELGQHALGGLLAHSAGAMLINGSTVNSYKRYWDAGQFAPSRINWGLDNKTCTVRLSAVGR LEYKLPDAAVNPYLSHAVILAACEDGMKNEIDPGAPTQGSSYDAPVDTRFPALPLTLGEAIEAFRADEVL TQALGPDLSSLLIDFHADEWARFCGYVTDWEREMYWSDAP*
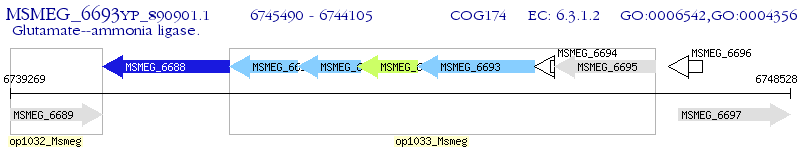
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6693 | - | - | 100% (461) | glutamine synthetase III |
| M. smegmatis MC2 155 | MSMEG_5374 | - | 3e-38 | 31.55% (393) | glutamate--ammonia ligase |
| M. smegmatis MC2 155 | MSMEG_4294 | glnA | 6e-37 | 27.45% (459) | glutamine synthetase, type I |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2246c | glnA2 | 5e-36 | 28.17% (465) | glutamine synthetase |
| M. gilvum PYR-GCK | Mflv_1300 | - | 1e-36 | 30.11% (455) | glutamate--ammonia ligase |
| M. tuberculosis H37Rv | Rv2222c | glnA2 | 6e-36 | 28.17% (465) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 3e-35 | 28.21% (475) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 4e-36 | 28.04% (460) | glutamine synthetase |
| M. marinum M | MMAR_3294 | glnA2 | 2e-36 | 27.33% (461) | glutamine synthetase GlnA2 |
| M. avium 104 | MAV_2244 | glnA | 5e-35 | 26.80% (459) | glutamine synthetase, type I |
| M. thermoresistible (build 8) | TH_3396 | - | 3e-35 | 27.17% (460) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_1335 | glnA2 | 9e-37 | 27.55% (461) | glutamine synthetase GlnA2 |
| M. vanbaalenii PYR-1 | Mvan_1850 | - | 0.0 | 92.62% (461) | glutamate--ammonia ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2246c|M.bovis_AF2122/97 --------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAI
Rv2222c|M.tuberculosis_H37Rv --------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAI
MMAR_3294|M.marinum_M --------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAI
MUL_1335|M.ulcerans_Agy99 --------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAI
MAV_2244|M.avium_104 --------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAI
MLBr_01631|M.leprae_Br4923 --------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKSVAI
TH_3396|M.thermoresistible__bu --------------MDRQKEFVLRSLEERDIRFVRLWFTDVLGFLKSVAI
MAB_1920|M.abscessus_ATCC_1997 --------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKSVSI
Mflv_1300|M.gilvum_PYR-GCK -----------------MPHDLAALAEQSGTRFILALFVDLRGKPCAKLV
MSMEG_6693|M.smegmatis_MC2_155 MTITPLDAHRQANATSPDLAAVLETIAERGVEFVYFQAVTITGRVVGKVA
Mvan_1850|M.vanbaalenii_PYR-1 MTITPLDTHREANAGSPELAAVLSTIAERGVEFVYFQAITVTGRVVGKVA
: : . .*: : * .
Mb2246c|M.bovis_AF2122/97 APAELEGAFEEGIGFDGSSIEGFARVS----------ESDTVAHPDPSTF
Rv2222c|M.tuberculosis_H37Rv APAELEGAFEEGIGFDGSSIEGFARVS----------ESDTVAHPDPSTF
MMAR_3294|M.marinum_M APAELEGAFEEGIGFDGSSIEGFARVS----------ESDTVAHPDPSTF
MUL_1335|M.ulcerans_Agy99 APAELEGAFEEGIGFDGSSIEGFARVS----------ESDTVAHPDPSTF
MAV_2244|M.avium_104 APAELEGAFEEGIGFDGSSIEGFSRVS----------ESDTVANPDPSTF
MLBr_01631|M.leprae_Br4923 APAELEGAFEEGIGFDGSSIEGFARVS----------ESDTVAHPDPSTF
TH_3396|M.thermoresistible__bu APAELEGAFEEGIGFDGSAIEGFARVS----------EADMVARPDPSTF
MAB_1920|M.abscessus_ATCC_1997 APAELEGAFEEGIGFDGSSIEGFARVS----------ESDTVAHPDPSTF
Mflv_1300|M.gilvum_PYR-GCK PVEAVDQLATEGVGFAGYAVGAMGQEPK---------DPDLMAIPDPESF
MSMEG_6693|M.smegmatis_MC2_155 PAKHFERLAVRGVQQHQTAVANLQGTREGVLLAGGVNAPEYTAIPDLETF
Mvan_1850|M.vanbaalenii_PYR-1 PARHFERLAIRGVQQHQTAIANLQGTREGVLLAGGVNAPEYTAIPDLETF
. .: .*: :: : .: * ** .:*
Mb2246c|M.bovis_AF2122/97 QVLPWATSSGHHHSARMFCDITMPDGSPSWADPRHVLR-RQLTKAGELGF
Rv2222c|M.tuberculosis_H37Rv QVLPWATSSGHHHSARMFCDITMPDGSPSWADPRHVLR-RQLTKAGELGF
MMAR_3294|M.marinum_M QVLPWVTDAGHHHSARIFCDITMPDGSPSWADPRHVLR-RQLTKANELGF
MUL_1335|M.ulcerans_Agy99 QVLPWVTDAGHHHSARIFCDITMPDGSPSWADPRHVLR-RQLIKANELGF
MAV_2244|M.avium_104 QVLPWASPNGHHHSARMFCDITMPDGSPSWADPRHVLR-RQLQKANDLGF
MLBr_01631|M.leprae_Br4923 QILPWATTAGHHHSARMFCDITMPDGSSSWADPRHVLR-RQLTKANDLGF
TH_3396|M.thermoresistible__bu QVLPWTGDSGKHYSARMFCDITMPDGSPSWADPRHVLR-RQLAKAGDQGF
MAB_1920|M.abscessus_ATCC_1997 QVLPWTTSAGKHHSARMFCDIKLPDGSPSWSDPRHVLR-RQLAKASDLGF
Mflv_1300|M.gilvum_PYR-GCK TPIPFIKEG----LAIVHCDPHVN-GKPWPYAPRVILK-SLIQRCADAGF
MSMEG_6693|M.smegmatis_MC2_155 AVLPWDTSF-----ARVFCRLYEPDHLAERAGAEFACDSRGLLRRLHAGF
Mvan_1850|M.vanbaalenii_PYR-1 AVLPWDTSF-----ARVFCRLYEPDHLAERAGAEFACDTRGLLRRMLSGF
:*: * :.* . .. : : **
Mb2246c|M.bovis_AF2122/97 SCYVHPEIEFFLLKPGPEDG---SVPVPVDNAGYFDQAVHDSALNFRRHA
Rv2222c|M.tuberculosis_H37Rv SCYVHPEIEFFLLKPGPEDG---SVPVPVDNAGYFDQAVHDSALNFRRHA
MMAR_3294|M.marinum_M SCYVHPEIEFFLLNPGPEDG---TEPVPIDNAGYFDQAVHVSASKFRRHA
MUL_1335|M.ulcerans_Agy99 SCYVHPEIEFFLLNPGPEDG---TEPVPIDNAGYFDQAVHVSASKFRRHV
MAV_2244|M.avium_104 SCYVHPEIEFFLLKPGPDDG---TPPVPVDTAGYFDQAIHDSASNFRRHA
MLBr_01631|M.leprae_Br4923 SCYVHPEIEFFLLKAGPEDGS-MPIPVPVDNAGYFDQAVHDSALNFRRHA
TH_3396|M.thermoresistible__bu TCYVHPEIEFFLLKPGPHDG---SPPVPADTSGYFDQSVHDTAPNFRRHA
MAB_1920|M.abscessus_ATCC_1997 SCYVHPEIEFFLLKNLPDDG---TAPIPADDAGYFDQAVHEAAPNFRRHA
Mflv_1300|M.gilvum_PYR-GCK EPWVGAEVEYFLLRRNDDGSVSVADAADTASQPCYDARGVTRMYDHLTSV
MSMEG_6693|M.smegmatis_MC2_155 TDRTGLELRTGCEPEMTWQGEGLEAQFRPGSSPAYHIEHLERNRPIVKKV
Mvan_1850|M.vanbaalenii_PYR-1 TDRTGLELRTGCEPEMTWQGDGLEAHFRPGSSPAYHIEHLERNRPIVKKV
. *:. . :. .
Mb2246c|M.bovis_AF2122/97 IDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMTFRYVIKEVA
Rv2222c|M.tuberculosis_H37Rv IDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMTFRYVIKEVA
MMAR_3294|M.marinum_M IDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMTFRYVIKEVA
MUL_1335|M.ulcerans_Agy99 IDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMTFRYVIKEVA
MAV_2244|M.avium_104 IEALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMTFRYVIKEVA
MLBr_01631|M.leprae_Br4923 IEALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADNVMTFRYVIKEVA
TH_3396|M.thermoresistible__bu IEALEQMGISVEFSHHEGAPGQQEIDLRYADALSMADNVMTFRYVVKQVA
MAB_1920|M.abscessus_ATCC_1997 IDALESMGISVEFSHHEGAPGQQEIDLRYADALSMADNVMTFRNVVKEVA
Mflv_1300|M.gilvum_PYR-GCK SAAMNQLGWSNYANDHEDGNGQFEQNFKFAEALTTADRVITLRYLLSMIA
MSMEG_6693|M.smegmatis_MC2_155 VEYAQALGLDMIEGDYED-EFQVELNFMYDRADLTADRLTTYRQICKQVA
Mvan_1850|M.vanbaalenii_PYR-1 VEYAQALGLDMIEGDYED-EFQVELNFMYDRADLTADRLTTYRQICKQVA
: :* . ..:*. * * :: : * **.: * * : . :*
Mb2246c|M.bovis_AF2122/97 LEEGARASFMPKPFGQHPGSAMHTHMSLFEGDVNAFHSADDP--LQLSEV
Rv2222c|M.tuberculosis_H37Rv LEEGARASFMPKPFGQHPGSAMHTHMSLFEGDVNAFHSADDP--LQLSEV
MMAR_3294|M.marinum_M LEEGVRATFMPKPFGQHAGSAMHTHMSLFEGDVNAFHSPDDP--LQLSEV
MUL_1335|M.ulcerans_Agy99 LEEGVRATFMPKPFGQHAGSAMHTHMSLFEGDVNAFHSPDDP--LQLSEV
MAV_2244|M.avium_104 IENGARASFMPKPFGQHPGSAMHTHMSLFEGDVNAFHSPDDP--LQLSDV
MLBr_01631|M.leprae_Br4923 IENGARASFMPKPFAQHPGSAMHTHMSLFEGDVNAFHSDDDP--LQLSDV
TH_3396|M.thermoresistible__bu ISDGVRASFMPKPFAEHPGSAMHTHMSLFEGDTNAFHSPDDP--LQLSDV
MAB_1920|M.abscessus_ATCC_1997 IIDGVHATFMPKPFSDHPGSAMHTHMSLFEGDTNAFHNPDDP--LQLSDV
Mflv_1300|M.gilvum_PYR-GCK DERGMIATFMPKPFGDKTGNGLHFHLSLTSAGTPVFPAEDDNRGLGLSDT
MSMEG_6693|M.smegmatis_MC2_155 RELGIQASFMPKPATGMMGNGCHHNFSLWRDGVNVLAEPGVT-ELHLSEL
Mvan_1850|M.vanbaalenii_PYR-1 RELGIHASFMPKPATGMMGNGCHHNFSLWRDDVNVLAEPGVT-ELHLSEV
* *:***** *.. * ::** .. .: . * **:
Mb2246c|M.bovis_AF2122/97 GKSFIAGILEHACEISAVTNQWVNSYKRLVQGGEAP-TAASWGAANRSAL
Rv2222c|M.tuberculosis_H37Rv GKSFIAGILEHACEISAVTNQWVNSYKRLVQGGEAP-TAASWGAANRSAL
MMAR_3294|M.marinum_M GKSFIAGILEHASEISAVTNQWVNSYKRLILGGEAP-TAASWGAANRSAL
MUL_1335|M.ulcerans_Agy99 GKSFIAGILEHASEISAVTNQWVNSYKRLILGGEAP-TAASWGAANRSAL
MAV_2244|M.avium_104 GKSFIAGILEHASEISAVTNQWVNSYKRLVGGGEAP-TAASWGAANRSAL
MLBr_01631|M.leprae_Br4923 GKSFIAGILEHASEISAVTNQWVNSYKRLVVGGEAP-TTASWGAANRSAL
TH_3396|M.thermoresistible__bu AKSFIAGILEHANEISAVTNQWVNSYKRLVHGGEAP-TAASWGAANRSAL
MAB_1920|M.abscessus_ATCC_1997 GKSFIAGILEHANEISAVTNQWVNSYKRLVQGGEAP-TAASWGAANRSAL
Mflv_1300|M.gilvum_PYR-GCK AYGFIGGILEHACALQAVVGPTVNSYKRTG-AVATL-SGASWAPRLPTYG
MSMEG_6693|M.smegmatis_MC2_155 GQHALGGLLAHSAGAMLINGSTVNSYKRYWDAGQFAPSRINWGLDNKTCT
Mvan_1850|M.vanbaalenii_PYR-1 GRHALGGLLAHSAGAMLINGSTVNSYKRYWDAGQFAPSRINWGLDNKTCT
. :.*:* *: : . ****** . : .*. :
Mb2246c|M.bovis_AF2122/97 VRVPMYTPHKTSSRRVEVRSPDSACNPYLTFAVLLAAGLRGVEKGYVLGP
Rv2222c|M.tuberculosis_H37Rv VRVPMYTPHKTSSRRVEVRSPDSACNPYLTFAVLLAAGLRGVEKGYVLGP
MMAR_3294|M.marinum_M VRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLLAAGLRGVEKGYVLGP
MUL_1335|M.ulcerans_Agy99 VRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLLAAGLRGVEKGYVLGP
MAV_2244|M.avium_104 VRVPMYTPHKTSSRRVEVRSPDSACNPYLTFAVLLAAGLRGVEKGYVLGP
MLBr_01631|M.leprae_Br4923 VRVPMYTPHKTSSRRVEVRSPDSACNPYLAFAVLLAAGLRGVEKGYVLGP
TH_3396|M.thermoresistible__bu VRVPMYTPHKASSRRVEVRSPDSACNPYLTFAVLLAAGLRGVEKGYVLGP
MAB_1920|M.abscessus_ATCC_1997 VRVPMYTPNKSSSRRIEVRSPDSACNPYLTYAVLLAAGLRGVEQGYTLGP
Mflv_1300|M.gilvum_PYR-GCK GNDRTHYIRVPDGDRIEMRGGDGSANPYLAIAAALGAGLDGIKRSTDPGA
MSMEG_6693|M.smegmatis_MC2_155 VR-------LSAVGRLEYKLPDAAVNPYLSHAVILAACEDGMKNEIDPGA
Mvan_1850|M.vanbaalenii_PYR-1 VR-------LSAVGRLEYKLPDAAVNPYLSHAVILAACEDGMKNATDPGK
. . *:* : *.: ****: *. *.* *::. *
Mb2246c|M.bovis_AF2122/97 QAEDNVWDLTPEERRAMGYRELPSSLDSALRAMEASELVA---EALGEHV
Rv2222c|M.tuberculosis_H37Rv QAEDNVWDLTPEERRAMGYRELPSSLDSALRAMEASELVA---EALGEHV
MMAR_3294|M.marinum_M QAEDNVWDLTPEERRTMGYRELPTSLDSALRAMESSELVA---EALGEHV
MUL_1335|M.ulcerans_Agy99 QAEDNVWDLTPEERRTMGYRELPTSLDSALRAMESSELVA---EALGEHV
MAV_2244|M.avium_104 QAEDNVWDLTPEERIAMGYRELPTSLDSALRAMESSELVA---ETLGEHV
MLBr_01631|M.leprae_Br4923 QAEDNVWDLTPEERLAMGYRELPTSLDSALGAMEVSEFVA---EALGEHV
TH_3396|M.thermoresistible__bu EAEDNVWTLTPEERRAMGFKELPSSLGQALAEMERSELVA---EALGEHV
MAB_1920|M.abscessus_ATCC_1997 EAEDDVWGLTRAERQAMGYKELPSTLENALIAMEGSELVA---EALGEHV
Mflv_1300|M.gilvum_PYR-GCK VGQG------------VSTQPLPATLLHAVEAFETNPVVTGVLDAAGEGV
MSMEG_6693|M.smegmatis_MC2_155 PTQGSSYDAPVDTR----FPALPLTLGEAIEAFRADEVLT---QALGPDL
Mvan_1850|M.vanbaalenii_PYR-1 PTQGSSYDAPVDGR----FPALPLTLGEAISAFRADEVLN---QAFGPDL
:. ** :* *: :. . .: :: * :
Mb2246c|M.bovis_AF2122/97 FDFFLRNKRTEWANYRSHVTPYELRTYLSL--
Rv2222c|M.tuberculosis_H37Rv FDFFLRNKRTEWANYRSHVTPYELRTYLSL--
MMAR_3294|M.marinum_M FDFFLRNKRREWATYRSHVTPYELSTYLSL--
MUL_1335|M.ulcerans_Agy99 FDFFLRNKRREWATYRSHVTPYELSTYLSL--
MAV_2244|M.avium_104 FDFFLRNKRTEWATYRSHVTPYELRTYLSL--
MLBr_01631|M.leprae_Br4923 FDFFLRNKRAEWANYRSHVSLYELKTYLSL--
TH_3396|M.thermoresistible__bu FDFFLRNKRAEWENYRSHVTPYELKNYLSL--
MAB_1920|M.abscessus_ATCC_1997 FDFFLRNKRAEWARYRSNVTPFELRAYLGL--
Mflv_1300|M.gilvum_PYR-GCK ASYFAAIKRDEFFAYHSSVSPWEIDYYLTAF-
MSMEG_6693|M.smegmatis_MC2_155 SSLLIDFHADEWARFCGYVTDWEREMYWSDAP
Mvan_1850|M.vanbaalenii_PYR-1 SALLVDYHADEWARFCGHVTEWEREMYWNDTP
: : *: : . *: :* *