For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPPLEQNTIGNGGDRAAVRSLTFDDITVTYVVDGVIALRPEVFFPAVPADAWTELVTAGGELLMSAGGLL VEIAGSTVLIDAGVGRMTETLSFGGVDCGSLIDVLGALGVPPGDIDVVAFTHLHFDHAGWAFTDGTKTFP NARYVVAADEIAPYGTAERNDDPTAPWDAITRLAGGEHDVVTVQDGEQVVPGVHAVVTGGHTPGHTSYVV TSAAGQRLVVFGDAFHTPAQLSHLHWLSSVDPDDGSVQGARLRLLTELDRPDTVGFAFHFGDQPFGRVVT DASGVRVWEPVASQLVAPPPRR
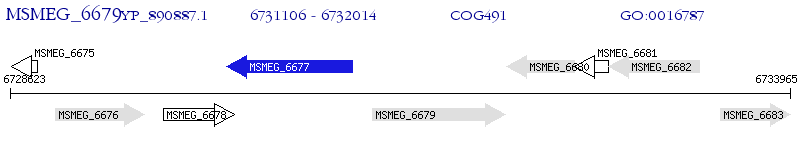
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6679 | - | - | 100% (302) | metallo-beta-lactamase family protein |
| M. smegmatis MC2 155 | MSMEG_3376 | - | 6e-19 | 29.41% (272) | beta-lactamase |
| M. smegmatis MC2 155 | MSMEG_0610 | - | 6e-19 | 26.85% (298) | beta-lactamase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0414c | - | 1e-05 | 29.19% (161) | beta lactamase like protein |
| M. gilvum PYR-GCK | Mflv_0212 | - | 3e-06 | 30.56% (216) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv0406c | - | 1e-05 | 29.19% (161) | beta lactamase like protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4947 | - | 2e-16 | 28.57% (259) | Beta-lactamase-like protein |
| M. marinum M | MMAR_3523 | - | 3e-06 | 27.78% (180) | hypothetical protein MMAR_3523 |
| M. avium 104 | MAV_4762 | - | 6e-05 | 31.54% (149) | metallo-beta-lactamase superfamily protein |
| M. thermoresistible (build 8) | TH_2871 | - | 1e-16 | 29.46% (241) | PUTATIVE beta-lactamase domain protein |
| M. ulcerans Agy99 | MUL_2815 | - | 3e-06 | 29.03% (186) | beta lactamase-like protein |
| M. vanbaalenii PYR-1 | Mvan_2949 | - | 2e-18 | 30.50% (282) | beta-lactamase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0414c|M.bovis_AF2122/97 -------------MVATRGTRL---AALALAPRLAGMAELVQITDKVHLA
Rv0406c|M.tuberculosis_H37Rv -------------MVATRGTRL---AALALAPRLAGMAELVQITDKVHLA
MAV_4762|M.avium_104 ------------------------------------MAELVQVTDAVHLA
MUL_2815|M.ulcerans_Agy99 -------------MPHQR-------------------TELVQVTDKVHLA
Mflv_0212|M.gilvum_PYR-GCK -----------------------------------MAPVVTAVTDRVHFV
MSMEG_6679|M.smegmatis_MC2_155 --MPPLEQNTIGNGGDRAAVRS---LTFDDITVTYVVDGVIALRPEVFFP
Mvan_2949|M.vanbaalenii_PYR-1 --MSLDGISRLGGQSTDELVPSRYVQQVGEIEVTVICDGVLPITASTLAT
MAB_4947|M.abscessus_ATCC_1997 MFSAAVRTFTVGDATVTQLTELDVWPIKPHHWYPALSEEQLVFARANYAP
TH_2871|M.thermoresistible__bu ----MIKTIEIGAVTVTRVMEWEGLFAPGTELIPDSDDRFWESERALLAP
MMAR_3523|M.marinum_M -----MKVHHLNCGTMNSPGAA----LLCHVLLVEAGNGLVLVDTGFGVQ
Mb0414c|M.bovis_AF2122/97 RGHAVNWV--------------------LVTDDTGVLLIDAG--------
Rv0406c|M.tuberculosis_H37Rv RGHAVNWV--------------------LVTDDTGVLLIDAG--------
MAV_4762|M.avium_104 RGDAVNWT--------------------LVADDTGVMLVDAG--------
MUL_2815|M.ulcerans_Agy99 QGPAVNWV--------------------LVTDETGVMLIDAG--------
Mflv_0212|M.gilvum_PYR-GCK RTELVNWT--------------------LVRDDDGVILIDAG--------
MSMEG_6679|M.smegmatis_MC2_155 AVPADAWT--------------------ELVTAGGELLMSAGG----LLV
Mvan_2949|M.vanbaalenii_PYR-1 NADPAELAGWLDDMFLPPDVLDWPLNVVVVRTGGQTILVDAG-----LGV
MAB_4947|M.abscessus_ATCC_1997 SAVSDDGSELIFAIHN-----------YVIELADTIIVVDTCSGNHKNRP
TH_2871|M.thermoresistible__bu DHWEPADRMVRACLQT-----------FVLQSEGRTILVDTGAGNHKERP
MMAR_3523|M.marinum_M DCLDPRRLG-------------------AFRHFLRPALLQNET-------
. ::.
Mb0414c|M.bovis_AF2122/97 -YPG---------------------DRAEVLASLNKLGYTPGDVRAIVLT
Rv0406c|M.tuberculosis_H37Rv -YPG---------------------DRAEVLASLNKLGYTPGDVRAIVLT
MAV_4762|M.avium_104 -YPG---------------------DREDVLGSLAELGYGPGDVRAIVLT
MUL_2815|M.ulcerans_Agy99 -YPG---------------------DRNNVLTSLRTLGYGPGDVRAILLT
Mflv_0212|M.gilvum_PYR-GCK -FPG---------------------HRDEVLTSLHGLGFGVGDVRAILLT
MSMEG_6679|M.smegmatis_MC2_155 EIAGSTVLIDAGVGRMTETLSFGGVDCGSLIDVLGALGVPPGDIDVVAFT
Mvan_2949|M.vanbaalenii_PYR-1 EFPDFP-------------------RAGQTVRRLEAAGIDPASVTDVVLT
MAB_4947|M.abscessus_ATCC_1997 LFADLH------------------MLNTDYLTRLEQSGYSPEAVDIVVNT
TH_2871|M.thermoresistible__bu YLPVFG------------------HLETDFLDRLAAAGFRPEDIDLVINT
MMAR_3523|M.marinum_M -----------------------------AVHQIERLGYRPSDVRHVVLT
: : * : : *
Mb0414c|M.bovis_AF2122/97 HAHIDHLGSAIWFAREHSTPVYCHAEEVGHAKREYRENASVFDVALRSWR
Rv0406c|M.tuberculosis_H37Rv HAHIDHLGSAIWFAREHSTPVYCHAEEVGHAKREYRENASVFDVALRSWR
MAV_4762|M.avium_104 HAHIDHLGTAIWLASEHGIPVYCHADEVGHAKREYLEQVSIVDVALRIWR
MUL_2815|M.ulcerans_Agy99 HAHIDHLGTAIWFAAQHGTPVYCDANEVGHAKREYHESASPLDVVLRSWR
Mflv_0212|M.gilvum_PYR-GCK HAHIDHFGTAIWFAATHGTPVYCHADEVAHSRRDHLEQASPIDIAKHAWQ
MSMEG_6679|M.smegmatis_MC2_155 HLHFDHAG----WAFTDGTKTFPNARYVVAA-----DEIAPYGTAERNDD
Mvan_2949|M.vanbaalenii_PYR-1 HLHMDHVG-GLLTDGLKARLRPDLRIHLAAAEAEFWAAPDFSQTAMPQPV
MAB_4947|M.abscessus_ATCC_1997 HLHLDHCGWNTRLVEGTWRPTFPAATYLFHHTELKYIRAQWESAPIDQWQ
TH_2871|M.thermoresistible__bu HVHVDHVGWNTYLHDREWVPTFPNARYVFSRRDFDFWNPLNGHERRGALV
MMAR_3523|M.marinum_M HFDFDHIGGLADFPDAQVHVTSAEARGAIHAPS--FRERVRYRSVQWAHG
* ..** *
Mb0414c|M.bovis_AF2122/97 PRVAVWGIHLLR-RGGLTGDG-IPTAQPLTAEAAAGLPGQPMAIFTPGHT
Rv0406c|M.tuberculosis_H37Rv PRVAVWGIHLLR-RGGLTGDG-IPTAQPLTAEAAAGLPGQPMAIFTPGHT
MAV_4762|M.avium_104 PRWAKWTAHVVR-SGGLIRDG-IPTAQPLTAEVAAGLPGRPTPVFSPGHT
MUL_2815|M.ulcerans_Agy99 PRTALWGIHLVF-RGGLRRAG-VPSTQALSAEVAAALPGHPIPIPTPGHT
Mflv_0212|M.gilvum_PYR-GCK PTWLKWSVSIIG-KGALTRAG-IASTAPLTDHIAAALPGSPVCIPTPGHT
MSMEG_6679|M.smegmatis_MC2_155 PTAPWDAITRLA-GGEHDVVT-VQDGEQVVPGVHAVVTGG----HTPGHT
Mvan_2949|M.vanbaalenii_PYR-1 PDVLRSVATRFL-DEYHGRLRPFETEYEVAPGVLISRTGG----HTPGHS
MAB_4947|M.abscessus_ATCC_1997 DNGGWVYEDSILPVLEHAPYGFVKSGQVLHSYGSTHIRALDAGGHTPGHV
TH_2871|M.thermoresistible__bu NQNMFEDSVLPVQQAGLADMW-EGDTHRIDANLTLELAPG----HTPGLS
MMAR_3523|M.marinum_M PKLVEHGHDGES----WRGFAAAKPLDAIADGVVLVPMPG----HTRGHA
: : *
Mb0414c|M.bovis_AF2122/97 SGHCSYVVDG-VLASGDALITGHPMLRHRGPQLLPAVFSHSQQNSIRSLA
Rv0406c|M.tuberculosis_H37Rv SGHCSYVVDG-VLASGDALITGHPMLRHRGPQLLPAVFSHSQQNSIRSLA
MAV_4762|M.avium_104 NGHCSYLIDG-VLVSGDALITGHPLLRHRGPQLLPAIFSYSQRDCIRTLS
MUL_2815|M.ulcerans_Agy99 SGHCSYLVDG-VLASGDALITGHPMLRHRGPQLLPAVFSKVQERCIRSLS
Mflv_0212|M.gilvum_PYR-GCK GGHCSYVVDG-VLVSGDALITGHPVASRRGPQLLPSVFNHDEDACRRSLT
MSMEG_6679|M.smegmatis_MC2_155 SYVVTSAAGQRLVVFGDAFHTPAQLSHLHWLSSVDPDDGSVQGARLRLLT
Mvan_2949|M.vanbaalenii_PYR-1 IVRVASGGEA-LTFAGDAVFAPG-FDNPEWHNGFEHDPEEAARVRVRLLR
MAB_4947|M.abscessus_ATCC_1997 AIEVRAADTG-LLIAGDALHHPMQAQFPDLPFYADADPAQAVATRRALLG
TH_2871|M.thermoresistible__bu VLKLESEGER-AVFVGDLLHSPVQILRPDWNSCFCEDPGRARASRRRVLS
MMAR_3523|M.marinum_M AVAVD-AGDRWILHCGDAFYHVGTLDGQSKVPFVLRSQEELFSFDRRQLR
** . *
Mb0414c|M.bovis_AF2122/97 ALALLETNILAPGHGELWHGPIRKATDEALERAQKSNHVFR-----
Rv0406c|M.tuberculosis_H37Rv ALALLETNILAPGHGELWHGPIRKATDEALERAQKSNHVFR-----
MAV_4762|M.avium_104 ALALLDTEVLLPGHGDVWRGPIKEATDAALALATGR----------
MUL_2815|M.ulcerans_Agy99 TLAQVEADTLAPGHGSLWRGPIRLATEEARARAQR-----------
Mflv_0212|M.gilvum_PYR-GCK TLGALDTEVILPGHGPLWRGPVRDAAAQAAQASVR-----------
MSMEG_6679|M.smegmatis_MC2_155 ELDRPDTVGFAFHFGDQPFGRVVTDASGVRVWEPVASQLVAPPPRR
Mvan_2949|M.vanbaalenii_PYR-1 ELAATGEALVATHLPFPSVCRVATAGDVFRCVPSVWDY--------
MAB_4947|M.abscessus_ATCC_1997 RCADENLKLLTAHFPAHSPLGVRRSGAAFTWDGSRAID--------
TH_2871|M.thermoresistible__bu WAADNNALLVPAHLGGDHAAEVVRAGDDFAVKGWAAFRSAGAV---
MMAR_3523|M.marinum_M DNQARLAELQRRQDPGLLIICAHDPSLYALAKDLS-----------