For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNEIVLGDVTVTRVMEYYGSVRLSPQTFFPESEPAAWQRNEHWLVPDFVDPDADVCVSAIQSWVLRSGGA TIVVDTGVGNNKERPDTPVWHRLQTDYLDNLAAAGVQPADVDYVINTHVHVDHVGWNTYLDNGHWAPTFP NATYLIPRADFEFWNPENRPSSSVDHRNLFMDSVAPIHAAGQALLWEDSFDINADLRLQAAPGHTPGSSI VTLESGSDRAVFVGDMLHTPLQFVEPDVNSCFCEDPAQARATRRRVLGWAADTRTLVVPAHLGGHGAAEL ARDGDTFAIKEWAPFERVTTHV
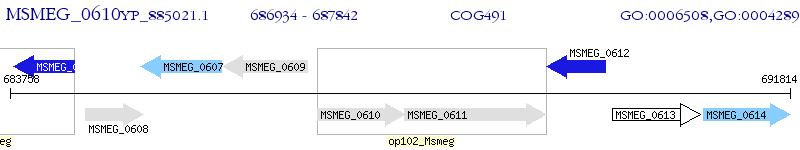
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0610 | - | - | 100% (302) | beta-lactamase |
| M. smegmatis MC2 155 | MSMEG_3376 | - | 2e-70 | 46.49% (299) | beta-lactamase |
| M. smegmatis MC2 155 | MSMEG_3427 | - | 3e-21 | 28.33% (293) | metal dependent hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4947 | - | 1e-38 | 35.02% (297) | Beta-lactamase-like protein |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2871 | - | 4e-92 | 55.74% (296) | PUTATIVE beta-lactamase domain protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5633 | - | 2e-64 | 43.09% (304) | beta-lactamase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0610|M.smegmatis_MC2_155 -----MNEIVLGDVTVTRVMEYYGSVRLSPQTFFPESEPAAWQRNEHWLV
TH_2871|M.thermoresistible__bu ----MIKTIEIGAVTVTRVMEWEGLFAPG-TELIPDSDDRFWESERALLA
Mvan_5633|M.vanbaalenii_PYR-1 ----MTGQVRLGRATLSRVVEVRAQMRTS---LFAQTPAAAWPANADLLE
MAB_4947|M.abscessus_ATCC_1997 MFSAAVRTFTVGDATVTQLTELDVWPIKP-HHWYPALSEEQLVFARANYA
. :* .*:::: * .
MSMEG_0610|M.smegmatis_MC2_155 PDFVDPDADVCVSAIQSWVLRSGGATIVVDTGVGNNKERPDTPVWHRLQT
TH_2871|M.thermoresistible__bu PDHWEPADRMVRACLQTFVLQSEGRTILVDTGAGNHKERPYLPVFGHLET
Mvan_5633|M.vanbaalenii_PYR-1 PLFWDRGADEWKIAIQTWVIRVDGLTVLVDTGVGNHRNRPHMPPLHQLDT
MAB_4947|M.abscessus_ATCC_1997 PSAVSDDGSELIFAIHNYVIELADTIIVVDTCSGNHKNRPLFADLHMLNT
* . .::.:*:. . ::*** **:::** . *:*
MSMEG_0610|M.smegmatis_MC2_155 DYLDNLAAAGVQPADVDYVINTHVHVDHVGWNTYLDNG--------HWAP
TH_2871|M.thermoresistible__bu DFLDRLAAAGFRPEDIDLVINTHVHVDHVGWNTYLHDR--------EWVP
Mvan_5633|M.vanbaalenii_PYR-1 PYLKALTDAGVSPEDVDVVVNTHLHTDHVGWNTRLSAASGAGTGGGAWTP
MAB_4947|M.abscessus_ATCC_1997 DYLTRLEQSGYSPEAVDIVVNTHLHLDHCGWNTRLVEG--------TWRP
:* * :* * :* *:***:* ** **** * * *
MSMEG_0610|M.smegmatis_MC2_155 TFPNATYLIPRADFEFWNPEN--RPSSSVDHRN--LFMDSVAPIHAAGQA
TH_2871|M.thermoresistible__bu TFPNARYVFSRRDFDFWNPLNGHERRGALVNQN--MFEDSVLPVQQAGLA
Mvan_5633|M.vanbaalenii_PYR-1 TFPNARYLMPELDYRHFSPDN---PDVADGMRI--VFADSVSPVEP--QM
MAB_4947|M.abscessus_ATCC_1997 TFPAATYLFHHTELKYIRAQWESAPIDQWQDNGGWVYEDSILPVLEHAPY
*** * *:: . : . . . :: **: *:
MSMEG_0610|M.smegmatis_MC2_155 LLWE-DSFDIN---ADLRLQAAPGHTPGSSIVTLESGSDRAVFVGDMLHT
TH_2871|M.thermoresistible__bu DMWEGDTHRID---ANLTLELAPGHTPGLSVLKLESEGERAVFVGDLLHS
Mvan_5633|M.vanbaalenii_PYR-1 ELFS-GDHRVS---DSLWLRPAPGHTPGSTMVWLDAG-VPAVFVGDLTHS
MAB_4947|M.abscessus_ATCC_1997 GFVKSGQVLHSYGSTHIRALDAGGHTPGHVAIEVRAADTGLLIAGDALHH
: . . . : * ***** : : : ::.** *
MSMEG_0610|M.smegmatis_MC2_155 PLQFVEPDVNSCFCEDPAQARATRRRVLGWAADTRTLVVPAHLGGHGAAE
TH_2871|M.thermoresistible__bu PVQILRPDWNSCFCEDPGRARASRRRVLSWAADNNALLVPAHLGGDHAAE
Mvan_5633|M.vanbaalenii_PYR-1 PIQIPRPDDPCAFDVDPMAAARSRHRILTEASRRRAAVIPAHYPGRGGAT
MAB_4947|M.abscessus_ATCC_1997 PMQAQFPDLPFYADADPAQAVATRRALLGRCADENLKLLTAHFPAHSPLG
*:* ** ** * :*: :* .: . ::.** .
MSMEG_0610|M.smegmatis_MC2_155 LARDGDTFAIKEWAPFERVTTHV
TH_2871|M.thermoresistible__bu VVRAGDDFAVKGWAAFRSAGAV-
Mvan_5633|M.vanbaalenii_PYR-1 VVARGNTFLVDQWLGFPEI----
MAB_4947|M.abscessus_ATCC_1997 VRRSGAAFTWDGSRAID------
: * * . :