For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPNAMLEGVRVLDFATLAAAPLAATYLGEFGAEVIKVEQPHHGDPIRTWGNQRDGIGLMWKSVSRNKKS ITLDLRSAEGQALAHRLVEHADVVVVNTRPQTLARWNMSYETLRRINDQIVMLHITGYGLSGPKAERPGF GTLGEAMSGFAHLTGEADGPPTLPPFMLADGVASLNAAYAVMMALYHRDVHGGDSQLIDINLIDPLARLL EQTLLGYDQLGLVPMRAGNRWDISAPRNTYQTADGKWLAMSGSSPALALRVFKAIGRADLVSDADFSDPQ RRLARAREVDAVVAEWVATKTLAEAMAVFDNAEVAAAPVYDIEGLVADEQLRHRGIFVAVEDSQLGRVTV QAPVPRFSDSDGRIDHLGPALGEHNDEIYGNLLGLLPAELDELRARGVL
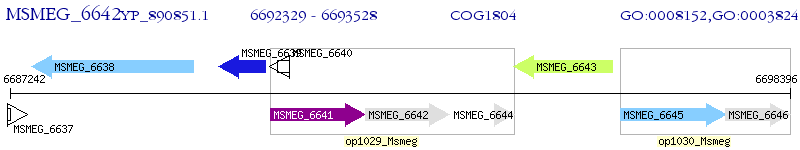
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6642 | - | - | 100% (399) | caib/baif family protein |
| M. smegmatis MC2 155 | MSMEG_6294 | - | 2e-91 | 43.51% (393) | caib/baif family protein |
| M. smegmatis MC2 155 | MSMEG_2073 | - | 1e-74 | 38.29% (397) | CAIB/BAIF family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1897 | - | 2e-32 | 28.04% (403) | hypothetical protein Mb1897 |
| M. gilvum PYR-GCK | Mflv_4465 | - | 4e-76 | 39.29% (397) | Formyl-CoA transferase |
| M. tuberculosis H37Rv | Rv1866 | - | 2e-32 | 28.04% (403) | hypothetical protein Rv1866 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4651c | - | 3e-75 | 39.14% (396) | putative L-carnitine dehydratase |
| M. marinum M | MMAR_1494 | - | 2e-75 | 38.56% (402) | hypothetical protein MMAR_1494 |
| M. avium 104 | MAV_0341 | - | 0.0 | 80.71% (394) | caib/baif family protein |
| M. thermoresistible (build 8) | TH_3905 | - | 1e-78 | 39.95% (403) | PUTATIVE Formyl-CoA transferase |
| M. ulcerans Agy99 | MUL_2446 | - | 3e-74 | 38.31% (402) | hypothetical protein MUL_2446 |
| M. vanbaalenii PYR-1 | Mvan_1895 | - | 7e-76 | 39.29% (397) | L-carnitine dehydratase/bile acid-inducible protein F |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6642|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0341|M.avium_104 --------------------------------------------------
MMAR_1494|M.marinum_M --------------------------------------------------
MUL_2446|M.ulcerans_Agy99 --------------------------------------------------
MAB_4651c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4465|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1895|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3905|M.thermoresistible__bu --------------------------------------------------
Mb1897|M.bovis_AF2122/97 MVTRLLADLGADVLKVQPPGGSPGRHVRPTLAGTSIGFAMHNANKRSAVL
Rv1866|M.tuberculosis_H37Rv MVTRLLADLGADVLKVEPPGGSPGRHVRPTLAGTSIGFAMHNANKRSAVL
MSMEG_6642|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0341|M.avium_104 --------------------------------------------------
MMAR_1494|M.marinum_M --------------------------------------------------
MUL_2446|M.ulcerans_Agy99 --------------------------------------------------
MAB_4651c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4465|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1895|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3905|M.thermoresistible__bu --------------------------------------------------
Mb1897|M.bovis_AF2122/97 NPLDESDRRRFLDLAASADIVVDCGLPGQAAAYGASCAELADRYRHLVAL
Rv1866|M.tuberculosis_H37Rv NPLDESDRRRFLDLAASADIVVDCGLPGQAAAYGASCAELADRYRHLVAL
MSMEG_6642|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0341|M.avium_104 --------------------------------------------------
MMAR_1494|M.marinum_M --------------------------------------------------
MUL_2446|M.ulcerans_Agy99 --------------------------------------------------
MAB_4651c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4465|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1895|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3905|M.thermoresistible__bu --------------------------------------------------
Mb1897|M.bovis_AF2122/97 SITDFGAAGPRSSWRATDPVLYAMSGALSRSGPTAGTPVLPPDGIASATA
Rv1866|M.tuberculosis_H37Rv SITDFGAAGPRSSWRATDPVLYAMSGALSRSGPTAGTPVLPPDGIASATA
MSMEG_6642|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0341|M.avium_104 --------------------------------------------------
MMAR_1494|M.marinum_M --------------------------------------------------
MUL_2446|M.ulcerans_Agy99 --------------------------------------------------
MAB_4651c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4465|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1895|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3905|M.thermoresistible__bu --------------------------------------------------
Mb1897|M.bovis_AF2122/97 AVQAAWAVLVAYFNRLRCGTGDYIDFSRFDAVVMALDPPFGAHGQVAAGI
Rv1866|M.tuberculosis_H37Rv AVQAAWAVLVAYFNRLRCGTGDYIDFSRFDAVVMALDPPFGAHGQVAAGI
MSMEG_6642|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0341|M.avium_104 --------------------------------------------------
MMAR_1494|M.marinum_M --------------------------------------------------
MUL_2446|M.ulcerans_Agy99 --------------------------------------------------
MAB_4651c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4465|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1895|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3905|M.thermoresistible__bu --------------------------------------------------
Mb1897|M.bovis_AF2122/97 RSTGRWRGRPKNQDAYPIYPCRDGYVRFCVMAPRQWRGLRRWLGEPEDFQ
Rv1866|M.tuberculosis_H37Rv RSTGRWRGRPKNQDAYPIYPCRDGYVRFCVMAPRQWRGLRRWLGEPEDFQ
MSMEG_6642|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0341|M.avium_104 --------------------------------------------------
MMAR_1494|M.marinum_M --------------------------------------------------
MUL_2446|M.ulcerans_Agy99 --------------------------------------------------
MAB_4651c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4465|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1895|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3905|M.thermoresistible__bu --------------------------------------------------
Mb1897|M.bovis_AF2122/97 DPKYDVIGARLAAWPQISVLVAKLCAEKTMKELVAAGQALGVPITAVLTP
Rv1866|M.tuberculosis_H37Rv DPKYDVIGARLAAWPQISVLVAKLCAEKTMKELVAAGQALGVPITAVLTP
MSMEG_6642|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0341|M.avium_104 --------------------------------------------------
MMAR_1494|M.marinum_M --------------------------------------------------
MUL_2446|M.ulcerans_Agy99 --------------------------------------------------
MAB_4651c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4465|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1895|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3905|M.thermoresistible__bu --------------------------------------------------
Mb1897|M.bovis_AF2122/97 SRILASEHFQAVGAITDAELVPGVRTGVPTGYFVVDGKRAGFRTPAPAAG
Rv1866|M.tuberculosis_H37Rv SRILASEHFQAVGAITDAELVPGVRTGVPTGYFVVDGKRAGFRTPAPAAG
MSMEG_6642|M.smegmatis_MC2_155 ------------------MTPNAMLEGVRVLDFATLAAAPLAATYLGEFG
MAV_0341|M.avium_104 -----------------------MLDGLRVLDLATLAAAPLVATYLGEFG
MMAR_1494|M.marinum_M ------------------MTEPGALDGIRVLELGTLIAGPFAGRLLGDMG
MUL_2446|M.ulcerans_Agy99 ------------------MTEPGALDGIRVLELGTLIAGPFAGRLLGDMG
MAB_4651c|M.abscessus_ATCC_199 --------------------MTGALDGIRVLELGTLIAGPFAGRLLGDMG
Mflv_4465|M.gilvum_PYR-GCK -------------------MTTGPLDGIRVIEVGTLISGPFAGRLLGDMG
Mvan_1895|M.vanbaalenii_PYR-1 -------------------MTTGPLDGIRVIEVGTLISGPFAGRLLGDMG
TH_3905|M.thermoresistible__bu ------------------VTVTGPLDGIRVLELGTLIAGPFAGRLLGDMG
Mb1897|M.bovis_AF2122/97 QDEPRWLADPAPVPPPSGRVGGYPFEGLRILDLGIIVAGGELSRLFGDLG
Rv1866|M.tuberculosis_H37Rv QDEPRWLADPAPVPPPSGRVGGYPFEGLRILDLGIIVAGGELSRLFGDLG
::*:*:::.. : :. . :*::*
MSMEG_6642|M.smegmatis_MC2_155 AEVIKVEQPHHGDPIRTWGN-QRDGIGLMWKSVSRNKKSITLDLRSAEGQ
MAV_0341|M.avium_104 ADVIKVEEPRHGDPIRGWGN-QRDGVGLMWKSISRNKKSITLDLRSADGQ
MMAR_1494|M.marinum_M ADVIKVEAPGAPDPLRTWGQAEVDGHHVFWTVHARNKRAITLDLRRPQGR
MUL_2446|M.ulcerans_Agy99 ADVIKVEAPGAPDPLRTWGQAEVDGHHVFWTVHARNKRAITLDLRRPQGR
MAB_4651c|M.abscessus_ATCC_199 AEVLKIEPPGAPDPLRTWGQAEVDGHHVFWTVHARNKKAITLDLRTEAGR
Mflv_4465|M.gilvum_PYR-GCK AEVIKVEPPGAPDPLRTWGQAELDGHHFFWTVHARNKKAVTLNLRVPEGR
Mvan_1895|M.vanbaalenii_PYR-1 AEVIKIEPPGAPDPLRTWGQAELDGHHFFWTVHARNKKAVTLNLRVPEGR
TH_3905|M.thermoresistible__bu AEVIKIELPGAPDPLRTWGQAELDGHHFFWTVHARNKKAITLNLREPAGR
Mb1897|M.bovis_AF2122/97 AEVIKVESADHPDGLRQTRVGDAMSESFAWTHR--NHLALGLDLRNSEGK
Rv1866|M.tuberculosis_H37Rv AEVIKVESADHPDGLRQTRVGDAMSESFAWTHR--NHLALGLDLRNSEGK
*:*:*:* . * :* : . . *. *: :: *:** *:
MSMEG_6642|M.smegmatis_MC2_155 ALAHRLVEHADVVVVNTRPQTLARWNMSYETLRRINDQIVMLHITGYGLS
MAV_0341|M.avium_104 QLVRRLVEHADVAIFNTRPQTLRKWGLDYESLRAVNERLVMLHITGYGLS
MMAR_1494|M.marinum_M ELFLELVDKSDIVVENFRPGTLEKWDLGYDVLRERNPGIILVRVSGYGQT
MUL_2446|M.ulcerans_Agy99 ELFLELVDKSDIVVENFRPGTLEKWDLGYDVLRERNPGIILVRVSGYGQT
MAB_4651c|M.abscessus_ATCC_199 ALFLDLVDKSDVIVENFRPGTLERWGLGYDVLAQRNKGIILVRVSGYGQT
Mflv_4465|M.gilvum_PYR-GCK DLFLDLVERSDVIVENFRPGTLEKWNLGYDVLRERNRGIILVRVSGYGQT
Mvan_1895|M.vanbaalenii_PYR-1 ELFLDLVERSDVIVENFRPGTLEKWGLGYDVLRERNRGIILVRVSGYGQT
TH_3905|M.thermoresistible__bu DVFLELVERSDIVVENFRPGTLERWDLGYDVLRSHNNRVILVRVSGYGQT
Mb1897|M.bovis_AF2122/97 AIFGRLVAESDAVFANFKPGTLTSLGFSYDVLHAFNPRIVLAGSSAFGNR
Rv1866|M.tuberculosis_H37Rv AIFGRLVAESDAVFANFKPGTLTSLGFSYDVLHAFNPRIVLAGSSAFGNR
: ** .:* . * :* ** .:.*: * * ::: :.:*
MSMEG_6642|M.smegmatis_MC2_155 GPKAERPGFGTLGEAMSGFAHLTGEADGPPTL-------PPFMLADGVAS
MAV_0341|M.avium_104 GPKSERPGFGTLGEAMSGFAHITGQAGAPPTL-------PPFMLADGVAS
MMAR_1494|M.marinum_M GPDAHKAGYASVAEAASGLRHLNGFPGGPPPR-------LALSLGDSLAG
MUL_2446|M.ulcerans_Agy99 GPDAHKAGYASVAEAASGLRHLNGFPGGPPPR-------LALSLGDSLAG
MAB_4651c|M.abscessus_ATCC_199 GPDAHKAGYASVAEAASGLRHLNGFPGGPPPR-------MALSIGDTLAG
Mflv_4465|M.gilvum_PYR-GCK GPDAHKAGYASVAEAASGLRHMNGFPGGPPPR-------LALSLGDSLAG
Mvan_1895|M.vanbaalenii_PYR-1 GPDAHKAGYASVAEAASGLRHMNGFPGGPPPR-------LALSLGDSLAG
TH_3905|M.thermoresistible__bu GPDAHKAGYASVAEAASGLRHLNGFPGGPPPR-------LALSLGDTLAG
Mb1897|M.bovis_AF2122/97 GPWSTRMGYGPLVRAATGVTRVWTSDEAQPDNSRHPFYDATTIFPDHVVG
Rv1866|M.tuberculosis_H37Rv GPWSTRMGYGPLVRAATGVTRVWTSDEAQPDNSRHPFYDATTIFPDHVVG
** : : *:..: .* :*. :: . * . : * :..
MSMEG_6642|M.smegmatis_MC2_155 LNAAYAVMMALYHRDVHGGDSQLIDINLIDPLARLLEQTLLGYDQLGLVP
MAV_0341|M.avium_104 LNAAYAVMMALYHRDVHGGPGQLIDVNLIDPLARLLEQTLLGYDQLGLVP
MMAR_1494|M.marinum_M MFGAQGALAALYRRSVTG-TGQVVDVALTESCLALQESTISDYDVGGVVR
MUL_2446|M.ulcerans_Agy99 MFGPQGALAALYRRSVTG-TGQVVDVALTESCLALQESTISDYDVGGVVR
MAB_4651c|M.abscessus_ATCC_199 MFAAQGALAALYRRTVTG-RGQVVDAALTESCLAVQESTIPDYDVGRVVR
Mflv_4465|M.gilvum_PYR-GCK MFAAQGALAALYRRSVTG-EGQIVDAALTESCLAVQESTIPDYDVGGVVR
Mvan_1895|M.vanbaalenii_PYR-1 MFAAQGALAALYRRSVTG-DGQVVDAALTESCLAVQESTIPDYDVGGVVR
TH_3905|M.thermoresistible__bu MFAAQGAMAALYRRTVTG-EGQVVDAALTEACLAVQESTIPDYDIGGVVR
Mb1897|M.bovis_AF2122/97 RVGALLALAALIHRDRTG-GGAHVHISQAEVVVNQLDTMFVAEAAR--AT
Rv1866|M.tuberculosis_H37Rv RVGALLALAALIHRDRTG-GGAHVHISQAEVVVNQLDTMFVAEAAR--AT
.. .: ** :* * . :. : : : .
MSMEG_6642|M.smegmatis_MC2_155 MRAGNRWDISAPRNTYQTADGKWLAMSGSSPALALRVFKAIGRADLVSDA
MAV_0341|M.avium_104 ERSGNRWDISAPRNTYQTADGRWLAMSGSSPALALRVFRAIGRDDLLGDP
MMAR_1494|M.marinum_M GPSGTRLEGIAPSNIYRSADESWVVIAANQDTVFTRLCQAMGCPELATDD
MUL_2446|M.ulcerans_Agy99 GPSGTRLKGIAPSNIYRSADESWVVIAANQDTVFTRLCQAMGCPELATDD
MAB_4651c|M.abscessus_ATCC_199 GPSGTRLEGIAPSNIYQSADGSWVVIAANQDTVFRRLCEAMGRPELSSDA
Mflv_4465|M.gilvum_PYR-GCK GPSGTRLEGIAPSNIYPTADGSWVVIAANQDTVFRRLCAAMGQPELATDE
Mvan_1895|M.vanbaalenii_PYR-1 GPSGTRLEGIAPSNIYPTADGSWVVIAANQDTVFRRLCAAMGQPELATDE
TH_3905|M.thermoresistible__bu GPSGTRLEGIAPSNIYRSADGSWVVIAANQDTVFRRLCAAMGRPELATDD
Mb1897|M.bovis_AF2122/97 DVAEIHPDTSVHAVYPCAGDDEWCVISIRSDDEWRRATSVFGQPELANDP
Rv1866|M.tuberculosis_H37Rv DVAEIHPDTSVHAVYPCAGDDEWCVISIRSDDEWRRATSVFGQPELANDP
: : . . :.* * .:: . * .:* :* *
MSMEG_6642|M.smegmatis_MC2_155 DFSDPQRRLARAREVDAVVAEWVATKTLAEAMAVFDNAEVAAAPVYDIEG
MAV_0341|M.avium_104 DFSDPQRRLARAREVDTLVADWVAGKTLSEAMAVFEAHEVAAAPVYDISD
MMAR_1494|M.marinum_M RFATHRARGRNQDELDKIIGAWAAQRQPDEIIETLSAAGVIAGPINTVAE
MUL_2446|M.ulcerans_Agy99 RFATHRARGRNQDELDKIIGAWAAQRQPDEIIETLSAAGVIAGPINTVAE
MAB_4651c|M.abscessus_ATCC_199 RFANHVARGRNQDDLDAIIAQWAAQRLPTDIIDVLSAAGVIAGPINTVAD
Mflv_4465|M.gilvum_PYR-GCK RFADHVARGRNQDEIDKIIGGWAAQRQPDEIISTLSEAGVISGPINTVAE
Mvan_1895|M.vanbaalenii_PYR-1 RFANHVARGRNQDELDKIIGAWAAERQPADIIATLSEAGVISGPINTVAE
TH_3905|M.thermoresistible__bu RFATHTARGRNQDELDKIIADWAAERAPGEIIDTLNAAGVIAGPINTVAE
Mb1897|M.bovis_AF2122/97 RFGASRSRVANRSELVAAVSAWTSTRTPVQAAGALQAAGVAAGPMNRPSD
Rv1866|M.tuberculosis_H37Rv RFGASRSRVANRSELVAAVSAWTSTRTPVQAAGALQAAGVAAGPMNRPSD
*. * . :: :. *.: : : .:. * :.*:
MSMEG_6642|M.smegmatis_MC2_155 LVADEQLRHRGIFVAVEDSQLGRVTVQAP--VPRFSDSDGRIDHLGP-AL
MAV_0341|M.avium_104 LVADEQLAHRGVFVSVDDQQLGPMTVQAP--VPRFSSASGTVEHLGP-RL
MMAR_1494|M.marinum_M VVKDRQLRARGMLVEHYDERIG-RNVLGPGIVPALSESPGQVRSAGPARP
MUL_2446|M.ulcerans_Agy99 VVKDRQLRARGMLVEHYDERIG-RNVLGPGIVPALSESPGQVRSAGPARP
MAB_4651c|M.abscessus_ATCC_199 VVQDPQLKAREMLVEHFDERLG-RSVLGPGVVPVLSESPGGVRNAGPARP
Mflv_4465|M.gilvum_PYR-GCK VVEDPQLQARGMIADHWDERIG-RNVKGPGVVPVLSETPGTIRSAGSARP
Mvan_1895|M.vanbaalenii_PYR-1 VVKDPQLQARGMIADHWDERIG-RNVKGPGVVPVLSETPGTIRSAGSSRP
TH_3905|M.thermoresistible__bu VVEDPQLRARGMIADHFDERIG-RNVKGPGVVPVLSESPGTIRNAGPARP
Mb1897|M.bovis_AF2122/97 ILEDPQLIERNLFRDMVHPLIA-RPLPAETGPAPFRHIPQAPQRPAP-LP
Rv1866|M.tuberculosis_H37Rv ILEDPQLIERNLFRDMVHPLIA-RPLPAETGPAPFRHIPQAPQRPAP-LP
:: * ** * :: . :. : . . : ..
MSMEG_6642|M.smegmatis_MC2_155 GEHNDEIYGNLLGLLPAELDELRARGVL-------
MAV_0341|M.avium_104 GEHNAEVYGELLGLTPDEIDQLRARGVV-------
MMAR_1494|M.marinum_M GQHNDDVYIGLLGKSIEELEQLRAEEVL-------
MUL_2446|M.ulcerans_Agy99 GQHNDDVYIGLLGKSIEELEQLRAVEVL-------
MAB_4651c|M.abscessus_ATCC_199 GQDNDDVYVGLLGKSAQDIERLRAEGVL-------
Mflv_4465|M.gilvum_PYR-GCK GQHNDEIYRDLLGRSAAELDKLQEEGVL-------
Mvan_1895|M.vanbaalenii_PYR-1 GEHNDEIYRGLLGKTDAELERLRAGGVL-------
TH_3905|M.thermoresistible__bu GQHNEEIYTGLLGKTAAELRQLRAQGVI-------
Mb1897|M.bovis_AF2122/97 GQDSVQICRKLLGMTADETERLINERVMFGPAVTA
Rv1866|M.tuberculosis_H37Rv GQDSVQICRKLLGMTADETERLINERVMFGPAVTA
*:.. :: *** : .* *: