For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFDTTSAQPPPLRGVRVVDGATIYAGPMIATLLGDFGADVIKVEHPHGDGLRQWEWRKDGESLWWALVGR NKRDISLSLSDPRGADVMRRLLADADVFIENFRPGTLEKWGLDPEDLIAANPKLVVVRVSGFGQTGPYRS RPGFGTIAEAMSGFADTNGAADGPPVLPQWPLADGVAALAGAFATMIALRHAEATGHGQVVDLSIYEPLF WILGAQTSAYDQLGTLPTRQGSRTGFNVPRECYRTRDGRWIALSGATTATAQRIMRAVGRPDLADAEWFS DISGRIEHRETIDEAIASWVGQRDNADVLAMFESVGATAGPVYTAADIVADPHFTERGSIVTVDHPTLGA VRMQGLIAQLSRTPGQIRHPGPAVGEHNDEILRTELAYPDDVVDALAEAGVIGRTLQPTSTKRVSS
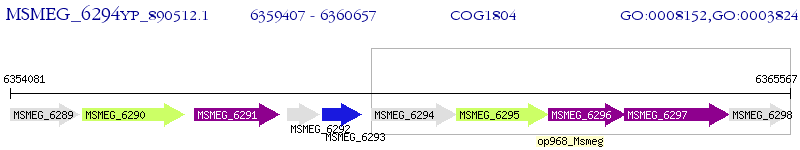
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6294 | - | - | 100% (416) | caib/baif family protein |
| M. smegmatis MC2 155 | MSMEG_6642 | - | 2e-91 | 43.51% (393) | caib/baif family protein |
| M. smegmatis MC2 155 | MSMEG_2073 | - | 6e-70 | 36.62% (396) | CAIB/BAIF family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3300 | - | 3e-39 | 29.31% (389) | hypothetical protein Mb3300 |
| M. gilvum PYR-GCK | Mflv_0637 | - | 1e-72 | 37.44% (398) | L-carnitine dehydratase/bile acid-inducible protein F |
| M. tuberculosis H37Rv | Rv3272 | - | 2e-39 | 29.31% (389) | hypothetical protein Rv3272 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4651c | - | 2e-64 | 34.94% (395) | putative L-carnitine dehydratase |
| M. marinum M | MMAR_1494 | - | 3e-69 | 37.57% (378) | hypothetical protein MMAR_1494 |
| M. avium 104 | MAV_0341 | - | 1e-95 | 45.04% (393) | caib/baif family protein |
| M. thermoresistible (build 8) | TH_3339 | - | 4e-93 | 42.57% (404) | PUTATIVE putative L-carnitine dehydratase/bile acid-inducible |
| M. ulcerans Agy99 | MUL_2446 | - | 7e-69 | 37.30% (378) | hypothetical protein MUL_2446 |
| M. vanbaalenii PYR-1 | Mvan_0528 | - | 2e-72 | 37.44% (398) | L-carnitine dehydratase/bile acid-inducible protein F |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6294|M.smegmatis_MC2_155 -----MFDTTSAQPPPLRGVRVVDGATIYAGPMIATLLGDFGADVIKVEH
MAV_0341|M.avium_104 -----MLD----------GLRVLDLATLAAAPLVATYLGEFGADVIKVEE
Mflv_0637|M.gilvum_PYR-GCK -----MSAN---MESLFDGLTILELGHVIAAPFAASLIGDFGAQVIKIED
Mvan_0528|M.vanbaalenii_PYR-1 -----MSAN---MESLFDGLTILELGHVIAAPFAASLIGDFGAQVIKIED
MMAR_1494|M.marinum_M ----------MTEPGALDGIRVLELGTLIAGPFAGRLLGDMGADVIKVEA
MUL_2446|M.ulcerans_Agy99 ----------MTEPGALDGIRVLELGTLIAGPFAGRLLGDMGADVIKVEA
MAB_4651c|M.abscessus_ATCC_199 ------------MTGALDGIRVLELGTLIAGPFAGRLLGDMGAEVLKIEP
TH_3339|M.thermoresistible__bu MTTEVETTATQPTTGPLHGIRVVEMGSLLAGPIVGQMMGDLGADVVKIED
Mb3300|M.bovis_AF2122/97 -------MPTSNPAKPLDGFRVLDFTQNVAGPLAGQVLVDLGAEVIKVEA
Rv3272|M.tuberculosis_H37Rv -------MPTSNPAKPLDGFRVLDFTQNVAGPLAGQVLVDLGAEVIKVEA
*. ::: *.*: . : ::**:*:*:*
MSMEG_6294|M.smegmatis_MC2_155 P-HGDGLRQWEWRK--DGESLWWALVGRNKRDISLSLSDPRGADVMRRLL
MAV_0341|M.avium_104 PRHGDPIRGWGNQR--DGVGLMWKSISRNKKSITLDLRSADGQQLVRRLV
Mflv_0637|M.gilvum_PYR-GCK PGSGDMLRRSGPRK--DGVPLWWKSAARNKSSVAVDLRLAEGQALVRKMV
Mvan_0528|M.vanbaalenii_PYR-1 PGSGDMLRRSGPRK--DGVPLWWKSAARNKSSVAVDLRLAEGQALVRKMV
MMAR_1494|M.marinum_M PGAPDPLRTWGQAEV-DGHHVFWTVHARNKRAITLDLRRPQGRELFLELV
MUL_2446|M.ulcerans_Agy99 PGAPDPLRTWGQAEV-DGHHVFWTVHARNKRAITLDLRRPQGRELFLELV
MAB_4651c|M.abscessus_ATCC_199 PGAPDPLRTWGQAEV-DGHHVFWTVHARNKKAITLDLRTEAGRALFLDLV
TH_3339|M.thermoresistible__bu PGKGDPLRQWGQNRP-KGQSLWFSIVGRNKRSVTINLREPRGQELVRQLV
Mb3300|M.bovis_AF2122/97 PGGEAARQITSVLPGRPPLATYFLPNNRGKKSVTVDLTTEQAKQQMLRLA
Rv3272|M.tuberculosis_H37Rv PGGEAARQITSVLPGRPPLATYFLPNNRGKKSVTVDLTTEQAKQQMLRLA
* : : *.* :::.* . . :
MSMEG_6294|M.smegmatis_MC2_155 ADADVFIENFRPGTLEKWGLDPEDLIAANPKLVVVRVSGFGQTGPYRSRP
MAV_0341|M.avium_104 EHADVAIFNTRPQTLRKWGLDYESLRAVNERLVMLHITGYGLSGPKSERP
Mflv_0637|M.gilvum_PYR-GCK ERADVVIENFRPGTLERWGLGWEELHAANPRLIMLRISGYGQIGPESAKP
Mvan_0528|M.vanbaalenii_PYR-1 ERADVVIENFRPGTLERWGLGWEELHAANPRLIMLRISGYGQIGPESAKP
MMAR_1494|M.marinum_M DKSDIVVENFRPGTLEKWDLGYDVLRERNPGIILVRVSGYGQTGPDAHKA
MUL_2446|M.ulcerans_Agy99 DKSDIVVENFRPGTLEKWDLGYDVLRERNPGIILVRVSGYGQTGPDAHKA
MAB_4651c|M.abscessus_ATCC_199 DKSDVIVENFRPGTLERWGLGYDVLAQRNKGIILVRVSGYGQTGPDAHKA
TH_3339|M.thermoresistible__bu AEADVLVENFRPGTMERWGLGYSDLAEINPRLVIVRVSGFGQDGPYASRP
Mb3300|M.bovis_AF2122/97 DTADVVLEAFRPGTMEKLGLGPDDLRSRNPNLIYARLTAYGGNGPHGSRP
Rv3272|M.tuberculosis_H37Rv DTADVVLEAFRPGTMEKLGLGPDDLRSRNPNLIYARLTAYGGNGPHGSRP
:*: : ** *:.: .*. . * * :: :::.:* ** :.
MSMEG_6294|M.smegmatis_MC2_155 GFGTIAEAMSGFADTNGAADGPPVLPQWPLADGVAALAGAFATMIALRHA
MAV_0341|M.avium_104 GFGTLGEAMSGFAHITGQAGAPPTLPPFMLADGVASLNAAYAVMMALYHR
Mflv_0637|M.gilvum_PYR-GCK GYGRVGEAMSGAVHITGHPDRPPTHFGFSLGDVTTGIMGAFAVAGALFKR
Mvan_0528|M.vanbaalenii_PYR-1 GYGRVGEAMSGAVHITGHPDRPPTHFGFSLGDVTTGIMGAFAVAGALFKR
MMAR_1494|M.marinum_M GYASVAEAASGLRHLNGFPGGPPPRLALSLGDSLAGMFGAQGALAALYRR
MUL_2446|M.ulcerans_Agy99 GYASVAEAASGLRHLNGFPGGPPPRLALSLGDSLAGMFGPQGALAALYRR
MAB_4651c|M.abscessus_ATCC_199 GYASVAEAASGLRHLNGFPGGPPPRMALSIGDTLAGMFAAQGALAALYRR
TH_3339|M.thermoresistible__bu GYGAIGEAMGGLRYVVGDPSTPPSRVGISIGDTLAALFATIGALAALRER
Mb3300|M.bovis_AF2122/97 GIDLVVAAEAGMTTGMPTPEGKPQIIPFQLVDNASGHVLAQAVLAALLHR
Rv3272|M.tuberculosis_H37Rv GIDLVVAAEAGMTTGMPTPEGKPQIIPFQLVDNASGHVLAQAVLAALLHR
* : * .* . * : * :. . .. ** .
MSMEG_6294|M.smegmatis_MC2_155 EATG---HGQVVDLSIYEPLFWILGAQTSAYDQLGTLPTRQGSRTGFNVP
MAV_0341|M.avium_104 DVHGG--PGQLIDVNLIDPLARLLEQTLLGYDQLGLVPERSGNRWDISAP
Mflv_0637|M.gilvum_PYR-GCK EAIGDRFDGECIDLALYESLFRGIDWQVILYDQLNFVAERQGNQFQVSPS
Mvan_0528|M.vanbaalenii_PYR-1 EAIGDRFDGECIDLALYESLFRGIDWQVILYDQLNFVAERQGNQFQVSPS
MMAR_1494|M.marinum_M SVTG---TGQVVDVALTESCLALQESTISDYDVGGVVRGPSGTRLEGIAP
MUL_2446|M.ulcerans_Agy99 SVTG---TGQVVDVALTESCLALQESTISDYDVGGVVRGPSGTRLKGIAP
MAB_4651c|M.abscessus_ATCC_199 TVTG---RGQVVDAALTESCLAVQESTIPDYDVGRVVRGPSGTRLEGIAP
TH_3339|M.thermoresistible__bu EISG---RGQMVDSAIYEAVLGVMESLVPEWTVSGYQRERTGAILPKIAP
Mb3300|M.bovis_AF2122/97 ERNG---VADVVQVAMYDVAVGLQANQLMMHLNRAASDQPKPEPAPKAKR
Rv3272|M.tuberculosis_H37Rv ERNG---VADVVQVAMYDVAVGLQANQLMMHLNRAASDQPKPEPAPKAKR
* .: :: : :
MSMEG_6294|M.smegmatis_MC2_155 ----------RECYRTRDGRWIALSGATTATAQRIMRAVGRPDLADAEWF
MAV_0341|M.avium_104 ----------RNTYQTADGRWLAMSGSSPALALRVFRAIGRDDLLGDPDF
Mflv_0637|M.gilvum_PYR-GCK PV--------SDTYLSGDGVWYTVATGTVRSVQSLLELLGGTTLRNDPKY
Mvan_0528|M.vanbaalenii_PYR-1 PV--------SDTYLSGDGVWYTVATGTVRSVQSLLELLGGTTLRNDPKY
MMAR_1494|M.marinum_M ----------SNIYRSADESWVVIAANQDTVFTRLCQAMGCPELATDDRF
MUL_2446|M.ulcerans_Agy99 ----------SNIYRSADESWVVIAANQDTVFTRLCQAMGCPELATDDRF
MAB_4651c|M.abscessus_ATCC_199 ----------SNIYQSADGSWVVIAANQDTVFRRLCEAMGRPELSSDARF
TH_3339|M.thermoresistible__bu ----------SNVYPTSDDTWILIAANQDTVFRRLAEAMGRPELAEDERY
Mb3300|M.bovis_AF2122/97 RKGVGFATQPSDAFRTADG-YIVISAYVPKHWQKLCYLIGRPDLVEDQRF
Rv3272|M.tuberculosis_H37Rv RKGVGFATQPSDAFRTADG-YIVISAYVPKHWQKLCYLIGRPDLVEDQRF
: : : * : :: : :* * :
MSMEG_6294|M.smegmatis_MC2_155 SDISGRIEHRETIDEAIASWVGQRDNADVLAMFESVGATAGPVYTAADIV
MAV_0341|M.avium_104 SDPQRRLARAREVDTLVADWVAGKTLSEAMAVFEAHEVAAAPVYDISDLV
Mflv_0637|M.gilvum_PYR-GCK ATQELQMLHREELGEMVRQWFAENNSDIVEKACASSGVVAARIFTPAEMF
Mvan_0528|M.vanbaalenii_PYR-1 ATQELQMLHREELGEMVRQWFAENNSDIVEKACASSGVVAARIFTPAEMF
MMAR_1494|M.marinum_M ATHRARGRNQDELDKIIGAWAAQRQPDEIIETLSAAGVIAGPINTVAEVV
MUL_2446|M.ulcerans_Agy99 ATHRARGRNQDELDKIIGAWAAQRQPDEIIETLSAAGVIAGPINTVAEVV
MAB_4651c|M.abscessus_ATCC_199 ANHVARGRNQDDLDAIIAQWAAQRLPTDIIDVLSAAGVIAGPINTVADVV
TH_3339|M.thermoresistible__bu ATHVARGEHQAELDEIVAEFTRRHTAAEVEALMHKHNVPVGKIYRPKEML
Mb3300|M.bovis_AF2122/97 AEQRSRSINYAELTAELELALASKTATEWVQLLQANGLMACLAHTWKQVV
Rv3272|M.tuberculosis_H37Rv AEQRSRSINYAELTAELELALASKTATEWVQLLQANGLMACLAHTWKQVV
: : . : : . . ::.
MSMEG_6294|M.smegmatis_MC2_155 ADPHFTERGSIVTVDHPTLG-AVRMQGLIAQLSRTPGQIRHPGPA-VGEH
MAV_0341|M.avium_104 ADEQLAHRGVFVSVDDQQLG-PMTVQAPVPRFSSASGTVEHLGPR-LGEH
Mflv_0637|M.gilvum_PYR-GCK SSPTFAARQSLVEVADQELG-PVRVTGVVPRLTNFPGSVRSTGPG-LGIH
Mvan_0528|M.vanbaalenii_PYR-1 SSPTFAARQSLVEVADQELG-PVRVTGVVPRLTNFPGSVRSTGPG-LGIH
MMAR_1494|M.marinum_M KDRQLRARGMLVEHYDERIGRNVLGPGIVPALSESPGQVRSAGPARPGQH
MUL_2446|M.ulcerans_Agy99 KDRQLRARGMLVEHYDERIGRNVLGPGIVPALSESPGQVRSAGPARPGQH
MAB_4651c|M.abscessus_ATCC_199 QDPQLKAREMLVEHFDERLGRSVLGPGVVPVLSESPGGVRNAGPARPGQD
TH_3339|M.thermoresistible__bu ADPQFRARESITSVDHPVLG-EIQMQNVFPRLSRTRGSIRWPGPG-LGEH
Mb3300|M.bovis_AF2122/97 DTPLFAESDLTLEVG--RGADTITVIRTPARYASFRAVVTDPPPT-AGEH
Rv3272|M.tuberculosis_H37Rv DTPLFAENDLTLEVG--RGADTITVIRTPARYASFRAVVTDPPPT-AGEH
: . : . : . : * * .
MSMEG_6294|M.smegmatis_MC2_155 NDEILRTELAYPDDVVDALAEAGVIGRTLQPTSTKRVSS
MAV_0341|M.avium_104 NAEVYGELLGLTPDEIDQLRARGVV--------------
Mflv_0637|M.gilvum_PYR-GCK GQEVLTEWLGMADSEYEALVSSGVVGAVDVDGEVPGH--
Mvan_0528|M.vanbaalenii_PYR-1 GQEVLTEWLGMADSEYEALVSSGVVGAVDVDGEVPGH--
MMAR_1494|M.marinum_M NDDVYIGLLGKSIEELEQLRAEEVL--------------
MUL_2446|M.ulcerans_Agy99 NDDVYIGLLGKSIEELEQLRAVEVL--------------
MAB_4651c|M.abscessus_ATCC_199 NDDVYVGLLGKSAQDIERLRAEGVL--------------
TH_3339|M.thermoresistible__bu TDEVLR-QLGLSDEELQKLHDAGVV--------------
Mb3300|M.bovis_AF2122/97 NAVFLARP-------------------------------
Rv3272|M.tuberculosis_H37Rv NAVFLARP-------------------------------
.