For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSTLRTRLSSRHRPGVLLAALLLTAGAAVACSAEDAAAGDAQAPGTASAIPDNTEVIAAIEADPALSAAL PPAIAQSKTINLGSNIQSAPNNFYAGDGKTPIGYEVDLAKAIAAKLGVQAKHQDMAFGSLITSLQSGRVD MTMAAMNDTKERQQQIDFVDYFSSGITIMIQKGNPENITGPDSLCGKNVAVVQGTSHQKFAAAQSQRCEQ EGKPAVNVTATDSDTQNQNQLRTGRVAAILNDLPSAVYISRTAGDGNAFEVVPGPPIEGGPYGIGFNKDN VALRDAVAKALGELMTDGTYETILQSWGVEQGALKEPAINGGS*
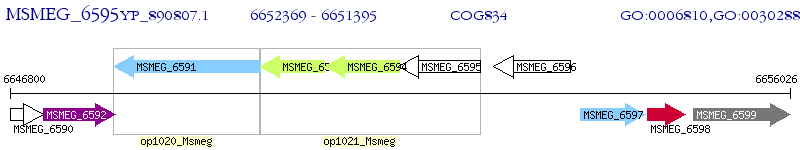
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6595 | - | - | 100% (324) | secreted protein |
| M. smegmatis MC2 155 | MSMEG_3235 | - | 2e-37 | 33.22% (304) | ABC-type amino acid transport system, secreted component |
| M. smegmatis MC2 155 | MSMEG_1612 | - | 8e-35 | 30.54% (298) | extracellular solute-binding protein, family protein 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0419c | glnH | 1e-11 | 25.09% (287) | glutamine-binding lipoprotein |
| M. gilvum PYR-GCK | Mflv_3583 | - | 5e-38 | 33.00% (300) | extracellular solute-binding protein |
| M. tuberculosis H37Rv | Rv0411c | glnH | 1e-11 | 25.09% (287) | glutamine-binding lipoprotein |
| M. leprae Br4923 | MLBr_00303 | glnH | 3e-10 | 24.20% (219) | putative glutamine-binding protein |
| M. abscessus ATCC 19977 | MAB_4236c | - | 3e-27 | 28.43% (306) | amino acid ABC transporter substrate-binding protein |
| M. marinum M | MMAR_0714 | glnH | 3e-11 | 26.67% (210) | glutamine-binding lipoprotein GlnH |
| M. avium 104 | MAV_4750 | - | 1e-12 | 26.28% (331) | extracellular solute-binding protein, family protein 3 |
| M. thermoresistible (build 8) | TH_0053 | - | 8e-23 | 29.79% (235) | ABC-type amino acid transport system, secreted component |
| M. ulcerans Agy99 | MUL_2809 | glnH | 2e-11 | 26.67% (210) | glutamine-binding lipoprotein GlnH |
| M. vanbaalenii PYR-1 | Mvan_2832 | - | 2e-36 | 33.45% (293) | extracellular solute-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0419c|M.bovis_AF2122/97 ------MTRRALLARAAAPLAPLALAMVLASCGHSETLGVEATPTLPLPT
Rv0411c|M.tuberculosis_H37Rv ------MTRRALLARAAAPLAPLALAMVLASCGHSETLGVEATPTLPLPT
MMAR_0714|M.marinum_M ------MRRLPGLRRAASALAVLALVITLDGCGHSEPLKVESTPTLPLPT
MUL_2809|M.ulcerans_Agy99 ------MRRLPGLRRAASALAVLALVITLDGCGHSEPLKVESTPTLPLPT
MAV_4750|M.avium_104 MAEAERVPMTAGLPLLRRACTALAAVLVAAGCGHTESLRVASVPTLPPPT
MLBr_00303|M.leprae_Br4923 --------MMTRQFKIQRACSVLATAMMLAACGHAELLTVASMPTLPPPT
Mflv_3583|M.gilvum_PYR-GCK ----MLGGIQGRQTRLWRVVAVFAATGAMALSGCSSSSESPSD-------
Mvan_2832|M.vanbaalenii_PYR-1 ----MVAGIQDRRTRIWRVAAVFAAVGALALSGCSSSSEEPVE-------
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_6595|M.smegmatis_MC2_155 ----MPSTLRTRLSSRHRPGVLLAALLLTAGAAVACSAEDAAAGDAQAPG
MAB_4236c|M.abscessus_ATCC_199 ----------------MRFLTVLVCTALTVLAGCAEGAAPGSS-------
Mb0419c|M.bovis_AF2122/97 PVGMEIMPPQPPLPPDSSSQDCDPTASLRPFATKAEADAAVADIRARGRL
Rv0411c|M.tuberculosis_H37Rv PVGMEIMPPQPPLPPDSSSQDCDPTASLRPFATKAEADAAVADIRARGRL
MMAR_0714|M.marinum_M PVGMEVMPPEPPLPADSSSQDCDPTASLRPFPTKAEADAAVADIRARGRL
MUL_2809|M.ulcerans_Agy99 PVGMEVMPPEPPLPADSSSQDCDPTASLRPFPTKAEADAAVADIRARGRL
MAV_4750|M.avium_104 PVGMEQLPPQPPLPPDGPDQNCDLTASLRPFPTKAEADAAVADIRARGRL
MLBr_00303|M.leprae_Br4923 PVGMEQVPPELLLPQDNTEEDCDPTASLRPFPTKAEADAAVAEIRARGRL
Mflv_3583|M.gilvum_PYR-GCK -----EGSPTAVAPAEKVD---------------AIANTVPESIKSTGTL
Mvan_2832|M.vanbaalenii_PYR-1 -----EGSPTAAAPAEKVD---------------AIANTVPEAIKSTGKL
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_6595|M.smegmatis_MC2_155 TASAIPDNTEVIAAIEADP---------------ALSAALPPAIAQSKTI
MAB_4236c|M.abscessus_ATCC_199 --SFNLSPDQDRVRVAKVD---------------AIAAEVPEGIRKRGTL
Mb0419c|M.bovis_AF2122/97 IVGLDIGSNL-FSFRDPITGEITGFDVDIAGEVARDIFGVPSHVEYRILS
Rv0411c|M.tuberculosis_H37Rv IVGLDIGSNL-FSFRDPITGEITGFDVDIAGEVARDIFGVPSHVEYRILS
MMAR_0714|M.marinum_M IVGLDIGSNL-FSFRDPITGEITGFDVDIASEVARDIFGGPTHVEYRILS
MUL_2809|M.ulcerans_Agy99 IVGLDIGSNL-FSFRDPITGEITGFDVDIASEVARDIFGGPTHVEYRILS
MAV_4750|M.avium_104 IVGLDIGSNL-FSFRDPITGEITGFDVDIAGEIARDIFGAPSHVEYRILS
MLBr_00303|M.leprae_Br4923 IVGLEIGSNL-FSFRDPITGEITGFDIDIAGEVARDIFGTPSHMEYRILS
Mflv_3583|M.gilvum_PYR-GCK VVGVNVPYAP-NEFKDPEGK-LVGFDVDLMNAIAGTLG---LTPEYKEAD
Mvan_2832|M.vanbaalenii_PYR-1 IVGVNIPYAP-NEFKDESGK-IVGFDVDLMNAIAGTLG---LTPEYREAD
TH_0053|M.thermoresistible__bu -----VPYKP-NEFKNAAGE-IVGFDVDLMNAVARTLG---LAPEYRELP
MSMEG_6595|M.smegmatis_MC2_155 NLGSNIQSAP-NNFYAGDGKTPIGYEVDLAKAIAAKLG---VQAKHQDMA
MAB_4236c|M.abscessus_ATCC_199 VVTGDANSSPPLRFYADDDKTMIGIETDIAYLVADILG---LRVDLRSAD
* * : *: :* : . :
Mb0419c|M.bovis_AF2122/97 AAERVTALQKSQVDIVVKTMSITCERRKLVNFSTVYLDANQRILAPRDSP
Rv0411c|M.tuberculosis_H37Rv AAERVTALQKSQVDIVVKTMSITCERRKLVNFSTVYLDANQRILAPRDSP
MMAR_0714|M.marinum_M AAERITALQSSQVDVVVKTMTITCERRKLVNFSTVYLDANQRILTSRDSP
MUL_2809|M.ulcerans_Agy99 AAERITALQSSQVDVVVKTMTITCERRKLVNFSTVYLDANQRILTSRDSP
MAV_4750|M.avium_104 SDERVTALQRGEVDVVVKTMTITCDRRKQVNFSTVYLDANQRILAPRDSP
MLBr_00303|M.leprae_Br4923 TAARITALQKSEVDIIVKTMTITCKRRKLVNFSTVYLDAKQRILAPRDSP
Mflv_3583|M.gilvum_PYR-GCK FAKIIPSIQGGTFDVGMSSFTDSKEREEQVDFVTYFSAGTAWAQPAGG--
Mvan_2832|M.vanbaalenii_PYR-1 FAKIIPSIQGGTFNVGMSSFTDSKEREQQVDFVTYFSAGTLWAQPAGG--
TH_0053|M.thermoresistible__bu FEAILPSVRAGDVHVGMASLTVTPERSAGADFVTYFQAGTLWAQPAGG--
MSMEG_6595|M.smegmatis_MC2_155 FGSLITSLQSGRVDMTMAAMNDTKERQQQIDFVDYFSSGITIMIQKGNPE
MAB_4236c|M.abscessus_ATCC_199 WAQNFVKVDSGEVDAFISNVTVTEERKEKFDFATYRLDNVTFEARRSLDW
. : . .. : .. : .* :*
Mb0419c|M.bovis_AF2122/97 -ITKVSDLSGKRVCVARGTTSLRRIREIA---------PPPVIVSVVNWA
Rv0411c|M.tuberculosis_H37Rv -ITKVSDLSGKRVCVARGTTSLRRIREIA---------PPPVIVSVVNWA
MMAR_0714|M.marinum_M -IFKVSDLSGKRVCVARGTTSLRRIREIA---------PPPVIVSVVNWA
MUL_2809|M.ulcerans_Agy99 -IFKVSDLSGKRVCVARGTTSLRRIREIA---------PPPVIVSVVNWA
MAV_4750|M.avium_104 -ITKVSDLSGKRVCVAKGTTSLHRIRQID---------PPPIVVSVVNWA
MLBr_00303|M.leprae_Br4923 -ITKPSDLSNKRVCMTRGTTSLHQVSEVV---------PPPIIVSAENWA
Mflv_3583|M.gilvum_PYR-GCK -DINPEDACGKKVAVQATTVQETDELPARSKKCVDEGKPAIEIIPFDSQD
Mvan_2832|M.vanbaalenii_PYR-1 -QIDPENACGKKVAVQATTVQETDELPARSKKCTDAGQPAIEIIAFDSQD
TH_0053|M.thermoresistible__bu -AVNPADACGLRVGVAYATIQESDEVPFKSEICTAAGKPPIDKVVYVSQD
MSMEG_6595|M.smegmatis_MC2_155 NITGPDSLCGKNVAVVQGTSHQK-FAAAQSQRCEQEGKPAVNVTATDSDT
MAB_4236c|M.abscessus_ATCC_199 KVKGARDIAGKTIGVGSGTNQEALLVRWNEEN-VAAGLPAADLKYFQQAT
. .. : : * *. .
Mb0419c|M.bovis_AF2122/97 DCLVALQQREIDAVSTDDTILAGLVEEDP----YLHIVGPDMADQPYGVG
Rv0411c|M.tuberculosis_H37Rv DCLVALQQREIDAVSTDDTILAGLVEEDP----YLHIVGPDMADQPYGVG
MMAR_0714|M.marinum_M DCLVALQQREIDAVSTDDTILAGLVEEDP----YLHIVGPNMANQPYGIG
MUL_2809|M.ulcerans_Agy99 DCLVALQQREIDAVSTDDTILAGLVEEDP----YLHIVGPNMANQPYGIG
MAV_4750|M.avium_104 DCLVAMQQREIDAVSTDDSILAGLVEEDP----YLHIVGPNMATQPYGIG
MLBr_00303|M.leprae_Br4923 DCLVAMQQREIDAISTDDAILAGLVEEDP----YLHIVGPNMATQPYGIG
Mflv_3583|M.gilvum_PYR-GCK AATNAVVLGQADAMSADSPVTLYAIKQTN---GKLEQAGETFDSAPYGWP
Mvan_2832|M.vanbaalenii_PYR-1 AATNAVVLGQADAMSADSPVTLYAIKQTN---GKLVQAGEVFDSAPYGWP
TH_0053|M.thermoresistible__bu DLTEALMNGEIDAMSADSPVTGFAVKTSG---GALQEAGEIFSPAPYGWP
MSMEG_6595|M.smegmatis_MC2_155 QNQNQLRTGRVAAILNDLPSAVYISRTAGDGNAFEVVPGPPIEGGPYGIG
MAB_4236c|M.abscessus_ATCC_199 GYYLALASGRIDAYVGPNPSAVFHSEATGQTKLVGTFSGAGEALQGEIAV
: . * . . *
Mb0419c|M.bovis_AF2122/97 INLDNTGLVRFVNGTLERIRNDGTWNTLYRKWLTVLGPAPAPPTPRYVD-
Rv0411c|M.tuberculosis_H37Rv INLDNTGLVRFVNGTLERIRNDGTWNTLYRKWLTVLGPAPAPPTPRYVD-
MMAR_0714|M.marinum_M INLDNTGLVRFVNGTLERIRNDGTWNTLYRKWLSVLGPAPAPPRPRYVD-
MUL_2809|M.ulcerans_Agy99 INLDNTGLVRFVNGTLERIRNDGTWNTLYRKWLSVLGPAPAPPRPRYVD-
MAV_4750|M.avium_104 INLNNTGLVRFVNGTLERIRRDGTWNTLYRKWLTVLGPAPAPPTPRYLD-
MLBr_00303|M.leprae_Br4923 INLNNTTLVRFVNGTLERIRRDGTWITLYRKWLTVLGPAPAPPTPRYVD-
Mflv_3583|M.gilvum_PYR-GCK VEK-GSPLAQSLLQALQHLIDNGKYQEIAANWGLEEGMIDKPVINGAIS-
Mvan_2832|M.vanbaalenii_PYR-1 VEK-GSPLAQSLLQALEHLIETGKYKEIAANWGLEEGMIDKPVINGAVS-
TH_0053|M.thermoresistible__bu VTR-GSPLAESLRQALEHLIATGEYRTIATMWGVEKGMIDRPEINAAAG-
MSMEG_6595|M.smegmatis_MC2_155 FNKDNVALRDAVAKALGELMTDGTYETILQSWGVEQGALKEPAINGGS--
MAB_4236c|M.abscessus_ATCC_199 LTKKDNGLVRAYADAINHAIADGTYRKVLQRWGLEDEAVAKSEINPRGLP
. . * :: . * : : * .
Mb0419c|M.bovis_AF2122/97 ---
Rv0411c|M.tuberculosis_H37Rv ---
MMAR_0714|M.marinum_M ---
MUL_2809|M.ulcerans_Agy99 ---
MAV_4750|M.avium_104 ---
MLBr_00303|M.leprae_Br4923 ---
Mflv_3583|M.gilvum_PYR-GCK ---
Mvan_2832|M.vanbaalenii_PYR-1 ---
TH_0053|M.thermoresistible__bu ---
MSMEG_6595|M.smegmatis_MC2_155 ---
MAB_4236c|M.abscessus_ATCC_199 RQG