For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKTALNVLAVGFAAVSLAGMVAGCSSSEGATPEPTGPAKVSPPAVLTADTLKICAPNDGTPPSVYHDESG DLVGSEVDLGKALAANLGLKPEFVQSAFAAVIPTLQAKQCDVIMAQLYIKPEREKVVDFVPYVYSGTGIA VSKAHPAAITGMDDSLCGKKVMVAVGTTAEVLSQEQSDKCTAAGKPPVDINRNNHADVSIQQVQNGQVDA YLDTAETIGYYETKTGAQIQMAGEPFGTIKIGAATLKGNTALHDAIQQALQAIEADGTYDKILDQWGQSE LSIQK
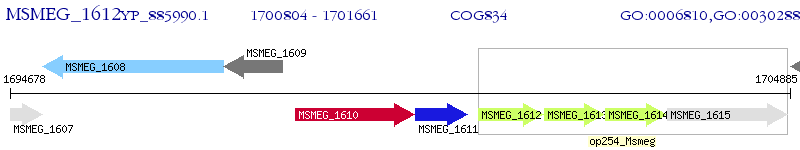
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1612 | - | - | 100% (285) | extracellular solute-binding protein, family protein 3 |
| M. smegmatis MC2 155 | MSMEG_6595 | - | 7e-35 | 30.54% (298) | secreted protein |
| M. smegmatis MC2 155 | MSMEG_0176 | - | 9e-32 | 31.12% (286) | glutamine ABC transporter periplasmic-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0419c | glnH | 2e-10 | 26.07% (211) | glutamine-binding lipoprotein |
| M. gilvum PYR-GCK | Mflv_3583 | - | 1e-30 | 33.33% (291) | extracellular solute-binding protein |
| M. tuberculosis H37Rv | Rv0411c | glnH | 2e-10 | 26.07% (211) | glutamine-binding lipoprotein |
| M. leprae Br4923 | MLBr_00303 | glnH | 9e-09 | 22.96% (257) | putative glutamine-binding protein |
| M. abscessus ATCC 19977 | MAB_4236c | - | 3e-17 | 26.82% (302) | amino acid ABC transporter substrate-binding protein |
| M. marinum M | MMAR_0061 | glnQ | 5e-12 | 25.09% (275) | ABC transporter permease protein GlnQ |
| M. avium 104 | MAV_0059 | - | 4e-11 | 26.55% (275) | amino acid ABC transporter, permease protein, |
| M. thermoresistible (build 8) | TH_0053 | - | 1e-27 | 35.16% (219) | ABC-type amino acid transport system, secreted component |
| M. ulcerans Agy99 | MUL_2809 | glnH | 1e-10 | 25.29% (257) | glutamine-binding lipoprotein GlnH |
| M. vanbaalenii PYR-1 | Mvan_2832 | - | 2e-30 | 32.77% (296) | extracellular solute-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3583|M.gilvum_PYR-GCK ---------------------------MLGGIQGRQTRLWRVVAVFAATG
Mvan_2832|M.vanbaalenii_PYR-1 ---------------------------MVAGIQDRRTRIWRVAAVFAAVG
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_1612|M.smegmatis_MC2_155 ----------------------------------MKTALNVLAVGFAAVS
MMAR_0061|M.marinum_M ---------------MLAVSATILIVLG--LALAAPALADHNQCAPAGVD
MAV_0059|M.avium_104 -----MGRRGTRRGKLLALTATVLMVCGSSALLAPPAAADRNQCAPAGPK
Mb0419c|M.bovis_AF2122/97 -MTRRALLARAAAPLAPLALAMVLASCGHSETLGVEATPTLPLPTPVGME
Rv0411c|M.tuberculosis_H37Rv -MTRRALLARAAAPLAPLALAMVLASCGHSETLGVEATPTLPLPTPVGME
MUL_2809|M.ulcerans_Agy99 -MRRLPGLRRAASALAVLALVITLDGCGHSEPLKVESTPTLPLPTPVGME
MLBr_00303|M.leprae_Br4923 MMTRQFKIQRACS---VLATAMMLAACGHAELLTVASMPTLPPPTPVGME
MAB_4236c|M.abscessus_ATCC_199 ------------------------------------MRFLTVLVCTALTV
Mflv_3583|M.gilvum_PYR-GCK AMALSGCSSSSESPSDEGSPTAVAPAEKVDAIANTVPESIKSTGTLVVGV
Mvan_2832|M.vanbaalenii_PYR-1 ALALSGCSSSSEEPVEEGSPTAAAPAEKVDAIANTVPEAIKSTGKLIVGV
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_1612|M.smegmatis_MC2_155 LAGMVAGCSSSEG----------ATPEPTGPAKVSPPAVLTADTLKICAP
MMAR_0061|M.marinum_M SAMVLPADLSSATTAGSDEDVQTTAGVVPLSSVNIGDLGLGTPGVLTAGT
MAV_0059|M.avium_104 SAAVLPENLTRGGSMS-QPDEHTTPTVEPLSSVRIDTLGLGTPGVLTVGT
Mb0419c|M.bovis_AF2122/97 IMPPQPPLPPDSSSQDCDPTASLRP-FATKAEADAAVADIRARGRLIVGL
Rv0411c|M.tuberculosis_H37Rv IMPPQPPLPPDSSSQDCDPTASLRP-FATKAEADAAVADIRARGRLIVGL
MUL_2809|M.ulcerans_Agy99 VMPPEPPLPADSSSQDCDPTASLRP-FPTKAEADAAVADIRARGRLIVGL
MLBr_00303|M.leprae_Br4923 QVPPELLLPQDNTEEDCDPTASLRP-FPTKAEADAAVAEIRARGRLIVGL
MAB_4236c|M.abscessus_ATCC_199 LAGCAEGAAPGSSSFNLSPDQDRVRVAKVDAIAAEVPEGIRKRGTLVVTG
Mflv_3583|M.gilvum_PYR-GCK NV-PYAPNEFKD-PEGKLVGFDVDLMNAIAGTLG---LTPEYKEADFAKI
Mvan_2832|M.vanbaalenii_PYR-1 NI-PYAPNEFKD-ESGKIVGFDVDLMNAIAGTLG---LTPEYREADFAKI
TH_0053|M.thermoresistible__bu -V-PYKPNEFKN-AAGEIVGFDVDLMNAVARTLG---LAPEYRELPFEAI
MSMEG_1612|M.smegmatis_MC2_155 ND-GTPPSVYHD-ESGDLVGSEVDLGKALAANLG---LKPEFVQSAFAAV
MMAR_0061|M.marinum_M LS-DAPPNTCIN-STGQFTGFDNELLRAIAAKLG---LQVRFVGTDFSGL
MAV_0059|M.avium_104 LS-GAPPNVCIT-PTGQYSGFDNQLLRAIAGKLG---LQVRFVGTDFAGL
Mb0419c|M.bovis_AF2122/97 DI-GSNLFSFRDPITGEITGFDVDIAGEVARDIFGVPSHVEYRILSAAER
Rv0411c|M.tuberculosis_H37Rv DI-GSNLFSFRDPITGEITGFDVDIAGEVARDIFGVPSHVEYRILSAAER
MUL_2809|M.ulcerans_Agy99 DI-GSNLFSFRDPITGEITGFDVDIASEVARDIFGGPTHVEYRILSAAER
MLBr_00303|M.leprae_Br4923 EI-GSNLFSFRDPITGEITGFDIDIAGEVARDIFGTPSHMEYRILSTAAR
MAB_4236c|M.abscessus_ATCC_199 DANSSPPLRFYADDDKTMIGIETDIAYLVADILG---LRVDLRSADWAQN
* : :: :* :
Mflv_3583|M.gilvum_PYR-GCK IPSIQGGTFDVGMSSFTDSKEREEQVDFVTYFSAGT----AWAQPAGGDI
Mvan_2832|M.vanbaalenii_PYR-1 IPSIQGGTFNVGMSSFTDSKEREQQVDFVTYFSAGT----LWAQPAGGQI
TH_0053|M.thermoresistible__bu LPSVRAGDVHVGMASLTVTPERSAGADFVTYFQAGT----LWAQPAGGAV
MSMEG_1612|M.smegmatis_MC2_155 IPTLQAKQCDVIMAQLYIKPEREKVVDFVPYVYSGTGIAVSKAHPAAITG
MMAR_0061|M.marinum_M LAQVAARRFDVGSASVKLTEARRRTVAFTNGYDFGY-YSLVVPPGSAITK
MAV_0059|M.avium_104 LAQVASRRFDVGSSSIKATDERRQTVAFTNGYDFGY-YALVVPPGSAIKE
Mb0419c|M.bovis_AF2122/97 VTALQKSQVDIVVKTMSITCERRKLVNFSTVYLDAN-QRILAPRDSPITK
Rv0411c|M.tuberculosis_H37Rv VTALQKSQVDIVVKTMSITCERRKLVNFSTVYLDAN-QRILAPRDSPITK
MUL_2809|M.ulcerans_Agy99 ITALQSSQVDVVVKTMTITCERRKLVNFSTVYLDAN-QRILTSRDSPIFK
MLBr_00303|M.leprae_Br4923 ITALQKSEVDIIVKTMTITCKRRKLVNFSTVYLDAK-QRILAPRDSPITK
MAB_4236c|M.abscessus_ATCC_199 FVKVDSGEVDAFISNVTVTEERKEKFDFATYRLDNVTFEARRSLDWKVKG
. : . . . * * .
Mflv_3583|M.gilvum_PYR-GCK NPEDACGKKVAVQATTVQETDELPARSKKCVDEGKPAIEIIPFDSQDAAT
Mvan_2832|M.vanbaalenii_PYR-1 DPENACGKKVAVQATTVQETDELPARSKKCTDAGQPAIEIIAFDSQDAAT
TH_0053|M.thermoresistible__bu NPADACGLRVGVAYATIQESDEVPFKSEICTAAGKPPIDKVVYVSQDDLT
MSMEG_1612|M.smegmatis_MC2_155 MDDSLCGKKVMVAVGTTAEVLSQ-EQSDKCTAAGKPPVDINRNNHADVSI
MMAR_0061|M.marinum_M FSDLAPGQRIGVVQGTV---------EESYVVNSLH-LQPVKYPDFATVY
MAV_0059|M.avium_104 FTDLAAGQRIGVVQGTV---------EESYVVDTLH-LQPVKYPDFATVY
Mb0419c|M.bovis_AF2122/97 VSDLS-GKRVCVARGTT---------SLRRIREIAPPPVIVSVVNWADCL
Rv0411c|M.tuberculosis_H37Rv VSDLS-GKRVCVARGTT---------SLRRIREIAPPPVIVSVVNWADCL
MUL_2809|M.ulcerans_Agy99 VSDLS-GKRVCVARGTT---------SLRRIREIAPPPVIVSVVNWADCL
MLBr_00303|M.leprae_Br4923 PSDLS-NKRVCMTRGTT---------SLHQVSEVVPPPIIVSAENWADCL
MAB_4236c|M.abscessus_ATCC_199 ARDIA-GKTIGVGSGTNQEALLVR-WNEENVAAGLPAADLKYFQQATGYY
. : : * .
Mflv_3583|M.gilvum_PYR-GCK NAVVLGQADAMSADSPVTLYAIKQTNGKLEQAGETFDSAPYG---WPVEK
Mvan_2832|M.vanbaalenii_PYR-1 NAVVLGQADAMSADSPVTLYAIKQTNGKLVQAGEVFDSAPYG---WPVEK
TH_0053|M.thermoresistible__bu EALMNGEIDAMSADSPVTGFAVKTSGGALQEAGEIFSPAPYG---WPVTR
MSMEG_1612|M.smegmatis_MC2_155 QQVQNGQVDAYLDTAETIGYYETKTGAQIQMAGEPFGTIKIG---AATLK
MMAR_0061|M.marinum_M GNLKTHQIDAWVAPGNQAEAAIRPGDPAVLVANTFGAGDFVA---YVVAR
MAV_0059|M.avium_104 ASLKTRQIDAWVAPAGQAATTIQPGDPAVVVANTISPGNFVA---YAVAK
Mb0419c|M.bovis_AF2122/97 VALQQREIDAVSTDDTILAGLVEE-DPYLHIVGPDMADQPYG---VGINL
Rv0411c|M.tuberculosis_H37Rv VALQQREIDAVSTDDTILAGLVEE-DPYLHIVGPDMADQPYG---VGINL
MUL_2809|M.ulcerans_Agy99 VALQQREIDAVSTDDTILAGLVEE-DPYLHIVGPNMANQPYG---IGINL
MLBr_00303|M.leprae_Br4923 VAMQQREIDAISTDDAILAGLVEE-DPYLHIVGPNMATQPYG---IGINL
MAB_4236c|M.abscessus_ATCC_199 LALASGRIDAYVGPNPSAVFHSEATGQTKLVGTFSGAGEALQGEIAVLTK
: . ** .
Mflv_3583|M.gilvum_PYR-GCK G-SPLAQSLLQALQHLIDNGKYQEIAANWGLEEGMIDKPVINGAIS----
Mvan_2832|M.vanbaalenii_PYR-1 G-SPLAQSLLQALEHLIETGKYKEIAANWGLEEGMIDKPVINGAVS----
TH_0053|M.thermoresistible__bu G-SPLAESLRQALEHLIATGEYRTIATMWGVEKGMIDRPEINAAAG----
MSMEG_1612|M.smegmatis_MC2_155 GNTALHDAIQQALQAIEADGTYDKILDQWGQSELSIQK------------
MMAR_0061|M.marinum_M DNRPLLDALNSGLDAVIADGTWSQLYSKWVPRTLPPGWKPGSRAVAAPTL
MAV_0059|M.avium_104 DNKPLVDALNSGLDAVIADGTWSNLYSQWVPRTLPPGWKPGSQAVAPPKL
Mb0419c|M.bovis_AF2122/97 DNTGLVRFVNGTLERIRNDGTWNTLYRKWLTVLGPAPAPPTPRYVD----
Rv0411c|M.tuberculosis_H37Rv DNTGLVRFVNGTLERIRNDGTWNTLYRKWLTVLGPAPAPPTPRYVD----
MUL_2809|M.ulcerans_Agy99 DNTGLVRFVNGTLERIRNDGTWNTLYRKWLSVLGPAPAPPRPRYVD----
MLBr_00303|M.leprae_Br4923 NNTTLVRFVNGTLERIRRDGTWITLYRKWLTVLGPAPAPPTPRYVD----
MAB_4236c|M.abscessus_ATCC_199 KDNGLVRAYADAINHAIADGTYRKVLQRWGLEDEAVAKSEINPRGLPRQG
* :: * : : *
Mflv_3583|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2832|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_1612|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0061|M.marinum_M PDFAAIAAAHRHKPVGPAAPKSALTQLRDSFFDWDMYRQAIPTLFKTGLP
MAV_0059|M.avium_104 PDFAAIAASQHHKAVGPAAPKSTLAQLRDSFFDWDMYRQAIPTLLRTGLP
Mb0419c|M.bovis_AF2122/97 --------------------------------------------------
Rv0411c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2809|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00303|M.leprae_Br4923 --------------------------------------------------
MAB_4236c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3583|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2832|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_1612|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0061|M.marinum_M NTLILTLSASIIGLALGMALAIAGISHQRWLRWPARVYTDIFRGLPEVVI
MAV_0059|M.avium_104 NTLILTFSASVIGLVLGMVLAVAGISHARWLRWPARVYTDVFRGLPEVVI
Mb0419c|M.bovis_AF2122/97 --------------------------------------------------
Rv0411c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2809|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00303|M.leprae_Br4923 --------------------------------------------------
MAB_4236c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3583|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2832|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_1612|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0061|M.marinum_M ILIIGLGVGPIVGGLTNNNPYPLGIAALGLMAGAYVGEILRSGIQSVDPG
MAV_0059|M.avium_104 ILLIGLGIGPIVGGLTNNNPYPLGIAALGMMAAAYIGEILRSGIQSVDPG
Mb0419c|M.bovis_AF2122/97 --------------------------------------------------
Rv0411c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2809|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00303|M.leprae_Br4923 --------------------------------------------------
MAB_4236c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3583|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2832|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_1612|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0061|M.marinum_M QLEASRALGFSYSAAMRLVVVPQGVRRVLPALVNQFIALLKASALVYFLG
MAV_0059|M.avium_104 QLEASRALGFSYATAMRLVVVPQGVRRVLPALVNQFIALLKASALVYFLG
Mb0419c|M.bovis_AF2122/97 --------------------------------------------------
Rv0411c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2809|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00303|M.leprae_Br4923 --------------------------------------------------
MAB_4236c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3583|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2832|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0053|M.thermoresistible__bu --------------------------------------------------
MSMEG_1612|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0061|M.marinum_M LIAQQRELFQVGRDLNAQTGNLSPLVAAGIFYLMLTVPLTHLVNYIDARL
MAV_0059|M.avium_104 LVADQRELFQVGRDLNAETGNLSPLVAAGACYLILTVPLTHLVNMIDARL
Mb0419c|M.bovis_AF2122/97 --------------------------------------------------
Rv0411c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2809|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00303|M.leprae_Br4923 --------------------------------------------------
MAB_4236c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3583|M.gilvum_PYR-GCK -------------------------
Mvan_2832|M.vanbaalenii_PYR-1 -------------------------
TH_0053|M.thermoresistible__bu -------------------------
MSMEG_1612|M.smegmatis_MC2_155 -------------------------
MMAR_0061|M.marinum_M RRGRRRLDPEDPPGVVSSTIGEEMT
MAV_0059|M.avium_104 RRGRAAVDPEEPAEMLT--FGQEIT
Mb0419c|M.bovis_AF2122/97 -------------------------
Rv0411c|M.tuberculosis_H37Rv -------------------------
MUL_2809|M.ulcerans_Agy99 -------------------------
MLBr_00303|M.leprae_Br4923 -------------------------
MAB_4236c|M.abscessus_ATCC_199 -------------------------