For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVAEPLPIVRLRHWGRWVGAAVIVAALVLLFIALSRAQIKWSSVPDFVVYRVMVVGLVNTVLLALVAQAA AIVIGVGIALLRRSANPVARWFAAAYIWLFRGLPVLLQILIWYNLALVIPVISIPLPMGGYLLQEPTNAL VSAFTAALLGLALNESAYMAEIVRAGLNSVDSGQVEAAKSIGMTPAQTLRRIVLPQAMRVIIPPTGNDFI DMLKGTSIASVIGVTELLHAANNISSNNLLVMETLFAAAIWYMVVVTLAGFGQHYLEQWFGGAERSAAAQ ATRALRAVPLKRSARV*
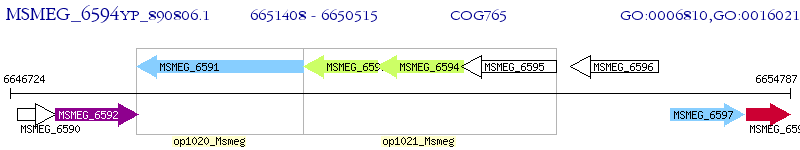
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6594 | - | - | 100% (297) | amino acid ABC transporter, permease protein, 3-TM region, His/Glu/Gln/Arg/opine |
| M. smegmatis MC2 155 | MSMEG_0177 | - | 6e-58 | 43.42% (281) | ABC polar amino acid family protein transporter, inner membrane |
| M. smegmatis MC2 155 | MSMEG_3236 | - | 1e-53 | 40.69% (290) | ABC-type amino acid transport system, permease component |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3582 | - | 7e-52 | 39.04% (292) | polar amino acid ABC transporter, inner membrane subunit |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4238c | - | 1e-45 | 35.14% (276) | amino acid ABC transporter permease |
| M. marinum M | MMAR_0061 | glnQ | 1e-20 | 30.77% (221) | ABC transporter permease protein GlnQ |
| M. avium 104 | MAV_0059 | - | 5e-20 | 28.83% (222) | amino acid ABC transporter, permease protein, |
| M. thermoresistible (build 8) | TH_2674 | - | 7e-25 | 37.17% (191) | PUTATIVE polar amino acid ABC transporter, inner membrane |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2833 | - | 1e-48 | 35.74% (291) | polar amino acid ABC transporter, inner membrane subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3582|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2833|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4238c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6594|M.smegmatis_MC2_155 --------------------------------------------------
TH_2674|M.thermoresistible__bu --------------------------------------------------
MMAR_0061|M.marinum_M ----------MLAVSATILIVLG--LALAAPALADHNQCAPAGVDSAMVL
MAV_0059|M.avium_104 MGRRGTRRGKLLALTATVLMVCGSSALLAPPAAADRNQCAPAGPKSAAVL
Mflv_3582|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2833|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4238c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6594|M.smegmatis_MC2_155 --------------------------------------------------
TH_2674|M.thermoresistible__bu --------------------------------------------------
MMAR_0061|M.marinum_M PADLSSATTAGSDEDVQTTAGVVPLSSVNIGDLGLGTPGVLTAGTLSDAP
MAV_0059|M.avium_104 PENLTRGGSMS-QPDEHTTPTVEPLSSVRIDTLGLGTPGVLTVGTLSGAP
Mflv_3582|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2833|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4238c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6594|M.smegmatis_MC2_155 --------------------------------------------------
TH_2674|M.thermoresistible__bu --------------------------------------------------
MMAR_0061|M.marinum_M PNTCINSTGQFTGFDNELLRAIAAKLGLQVRFVGTDFSGLLAQVAARRFD
MAV_0059|M.avium_104 PNVCITPTGQYSGFDNQLLRAIAGKLGLQVRFVGTDFAGLLAQVASRRFD
Mflv_3582|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2833|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4238c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6594|M.smegmatis_MC2_155 --------------------------------------------------
TH_2674|M.thermoresistible__bu --------------------------------------------------
MMAR_0061|M.marinum_M VGSASVKLTEARRRTVAFTNGYDFGYYSLVVPPGSAITKFSDLAPGQRIG
MAV_0059|M.avium_104 VGSSSIKATDERRQTVAFTNGYDFGYYALVVPPGSAIKEFTDLAAGQRIG
Mflv_3582|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2833|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4238c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6594|M.smegmatis_MC2_155 --------------------------------------------------
TH_2674|M.thermoresistible__bu --------------------------------------------------
MMAR_0061|M.marinum_M VVQGTVEESYVVNSLHLQPVKYPDFATVYGNLKTHQIDAWVAPGNQAEAA
MAV_0059|M.avium_104 VVQGTVEESYVVDTLHLQPVKYPDFATVYASLKTRQIDAWVAPAGQAATT
Mflv_3582|M.gilvum_PYR-GCK -------------------------------MTNEAGSPPTSGTPQSIDA
Mvan_2833|M.vanbaalenii_PYR-1 -------------------------------MT-DVGAPPPSSSPQSIDA
MAB_4238c|M.abscessus_ATCC_199 -------------------------------MTSAPAAPPRARVADEAAE
MSMEG_6594|M.smegmatis_MC2_155 -----------------------------------------MAVAEPLPI
TH_2674|M.thermoresistible__bu --------------------------------------------------
MMAR_0061|M.marinum_M IRPGDPAVLVANTFGAGDFVAYVVARDNRPLLDALNSGLDAVIADGTWSQ
MAV_0059|M.avium_104 IQPGDPAVVVANTISPGNFVAYAVAKDNKPLVDALNSGLDAVIADGTWSN
Mflv_3582|M.gilvum_PYR-GCK VPLRHPWR-----WVAAAVIIVLVGLFLWGAATNDAYGWST---------
Mvan_2833|M.vanbaalenii_PYR-1 IPLKHPWR-----WLAAAAIIVLVGLFLYGAATNPAYGWST---------
MAB_4238c|M.abscessus_ATCC_199 PVLRVRRRPRPGNWILSAVALVFVAMFVHGLVRNPGWDWPT---------
MSMEG_6594|M.smegmatis_MC2_155 VRLRHWGR-----WVGAAVIVAALVLLFIALSR-AQIKWSS---------
TH_2674|M.thermoresistible__bu --------------------------------------VQV---------
MMAR_0061|M.marinum_M LYSKWVPRTLPPGWKPGSRAVAAPTLPDFAAIAAAHRHKPVGPAAPKSAL
MAV_0059|M.avium_104 LYSQWVPRTLPPGWKPGSQAVAPPKLPDFAAIAASQHHKAVGPAAPKSTL
Mflv_3582|M.gilvum_PYR-GCK ------YAEYLFNERILLGVFNT-----IQLTVYSMVIGIVLGVILTIMR
Mvan_2833|M.vanbaalenii_PYR-1 ------FGEYFFHERILLGVFNT-----LQLTIYSMIIGIVLGVFLTVMR
MAB_4238c|M.abscessus_ATCC_199 ------FARYFTAKSVLAALWLT-----LRLTFYGTVLGFLLGVALAAAR
MSMEG_6594|M.smegmatis_MC2_155 ------VPDFVVYRVMVVGLVNT-----VLLALVAQAAAIVIGVGIALLR
TH_2674|M.thermoresistible__bu ------FSEYSG--QIFQAFWTT-----IQLTVLSAIGALVLGTVLAAMR
MMAR_0061|M.marinum_M TQLRDSFFDWDMYRQAIPTLFKTGLPNTLILTLSASIIGLALGMALAIAG
MAV_0059|M.avium_104 AQLRDSFFDWDMYRQAIPTLLRTGLPNTLILTFSASVIGLVLGMVLAVAG
: . . * : *:. . .: :* ::
Mflv_3582|M.gilvum_PYR-GCK LSSNPVLSSVAWVFLWIFRGTPVYVQLAFWGLMPTIYQNIQLGVPFGPSL
Mvan_2833|M.vanbaalenii_PYR-1 LSDNPVLSSVAWVFLWIFRGTPIYVQLAFWGLFPTIYQNIQLGVPFGPSF
MAB_4238c|M.abscessus_ATCC_199 LSKNPVLQAISWTYVWIFRSIPLIVQLLFWFNIAYLYDTIGFGIPFGPAL
MSMEG_6594|M.smegmatis_MC2_155 RSANPVARWFAAAYIWLFRGLPVLLQILIWYNLALVIPVISIPLPMGGYL
TH_2674|M.thermoresistible__bu LAPVPVLNWLGAGYVNVVRNTPLTLIILFCAFGLGQTLSITLADRGSPTF
MMAR_0061|M.marinum_M ISHQRWLRWPARVYTDIFRGLPEVVIILIIG--------LGVGPIVGGLT
MAV_0059|M.avium_104 ISHARWLRWPARVYTDVFRGLPEVVIILLIG--------LGIGPIVGGLT
: . : :.*. * : : : : . .
Mflv_3582|M.gilvum_PYR-GCK LHFDLQDLSFPFALAVIGLALNEAAYMAEIVRAGITSVPEGQLEASTALG
Mvan_2833|M.vanbaalenii_PYR-1 FHLDLQDLSIPFVLAVIGLGLNEAAYMAEIVRAGIMSVPEGQMEASTALG
MAB_4238c|M.abscessus_ATCC_199 FTVDTNELLSGLTAAVIGLTLHQGAYFAEIIRGGILSVDEGQLEAAAALG
MSMEG_6594|M.smegmatis_MC2_155 LQEPTNALVSAFTAALLGLALNESAYMAEIVRAGLNSVDSGQVEAAKSIG
TH_2674|M.thermoresistible__bu LQD------SNFRLAVLGFTVYTAAFVCETIRAGVNTVPVGQVEAARALG
MMAR_0061|M.marinum_M NNN-------PYPLGIAALGLMAGAYVGEILRSGIQSVDPGQLEASRALG
MAV_0059|M.avium_104 NNN-------PYPLGIAALGMMAAAYIGEILRSGIQSVDPGQLEASRALG
.: .: : .*:. * :*.*: :* **:**: ::*
Mflv_3582|M.gilvum_PYR-GCK MSWGRAMRRTVLPQAMRVIIPPTGNEVISLLKTTSLVTAVPYAFDLYSIA
Mvan_2833|M.vanbaalenii_PYR-1 MSWGKAMRRTVLPQAMRVIIPPTGNELISLLKTTSLVTAVPYAFDVYSIA
MAB_4238c|M.abscessus_ATCC_199 IPRRRQFFRIVLPQAMRSVMPNAANEVISLVKGTSIVSTMAIADLFYQVQ
MSMEG_6594|M.smegmatis_MC2_155 MTPAQTLRRIVLPQAMRVIIPPTGNDFIDMLKGTSIASVIGVTELLHAAN
TH_2674|M.thermoresistible__bu FTFGQNLRIIVLPQAFRAVIIPLGTVLIALTKNTTIASAIGVAEAALLMK
MMAR_0061|M.marinum_M FSYSAAMRLVVVPQGVRRVLPALVNQFIALLKASALVYFLGLIAQQRELF
MAV_0059|M.avium_104 FSYATAMRLVVVPQGVRRVLPALVNQFIALLKASALVYFLGLVADQRELF
:. : *:**..* :: . .* : * :::. :
Mflv_3582|M.gilvum_PYR-GCK TREIAARIFE-PVPLLLVAATWYLVITSILMVGQFYLERYFSRGVSRQLT
Mvan_2833|M.vanbaalenii_PYR-1 TREIAARIFE-PVPLLLVAAAWYLVITSILMVGQFYLERYFSRGASRKLT
MAB_4238c|M.abscessus_ATCC_199 V--IFGRNGR-VVPLLMVATAWYIVITSVLTVVQYYIERHYARGAHR---
MSMEG_6594|M.smegmatis_MC2_155 N--ISSNNLL-VMETLFAAAIWYMVVVTLAGFGQHYLEQWFG-GAERSAA
TH_2674|M.thermoresistible__bu EMIENTSAVL-IVGAIFAVG--FLILTLPLGLLSGRLSNRLAVRR-----
MMAR_0061|M.marinum_M QVGRDLNAQTGNLSPLVAAGIFYLMLTVPLTHLVNYIDARLRRGRRRLDP
MAV_0059|M.avium_104 QVGRDLNAETGNLSPLVAAGACYLILTVPLTHLVNMIDARLRRGRAAVDP
: :... ::::. :.
Mflv_3582|M.gilvum_PYR-GCK SKQLEALAKAQLGSAVP-
Mvan_2833|M.vanbaalenii_PYR-1 SKQLEALALAQLPQVK--
MAB_4238c|M.abscessus_ATCC_199 ------------------
MSMEG_6594|M.smegmatis_MC2_155 AQATRALRAVPLKRSARV
TH_2674|M.thermoresistible__bu ------------------
MMAR_0061|M.marinum_M EDPPGVVSSTIGEEMT--
MAV_0059|M.avium_104 EEPAEMLT--FGQEIT--