For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LGNSIAVLLALCAAIFMAVGIVVRQRATLDVPAGKGVSTVMLRTLLRRKLWWAGTISAVMGYGFQALALG FGSLLLVQPVLVSALLFALPLSARLARRTVSRADWMWALLLTGALTVFVLLARTSTGTYSVSVSTTVTVA VVCTSVVALCVVLATRSSNWRRAVLLAAAVGVMFGVVAVLTKIVMHLVADGDALATFTTPALYLVVVLGV LATLLQQSAFHAGSLQTSVPTMLVLEPVVAVLLGSVVLGEHLSIAGWQPVALTLAIAAMIAATIALGRDE GAYEETLEAEMEAKTVTG*
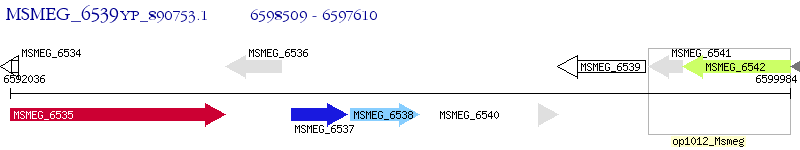
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6539 | - | - | 100% (299) | hypothetical protein MSMEG_6539 |
| M. smegmatis MC2 155 | MSMEG_2732 | - | 7e-57 | 46.32% (285) | hypothetical protein MSMEG_2732 |
| M. smegmatis MC2 155 | MSMEG_1505 | - | 2e-05 | 26.20% (229) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2748c | - | 4e-47 | 40.14% (289) | hypothetical protein Mb2748c |
| M. gilvum PYR-GCK | Mflv_1077 | - | 2e-99 | 67.28% (272) | hypothetical protein Mflv_1077 |
| M. tuberculosis H37Rv | Rv2729c | - | 6e-48 | 40.83% (289) | hypothetical protein Rv2729c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2553 | - | 6e-08 | 24.32% (259) | putative integral membrane protein |
| M. marinum M | MMAR_1984 | - | 9e-46 | 39.02% (287) | hypothetical protein MMAR_1984 |
| M. avium 104 | MAV_1660 | - | 7e-49 | 40.97% (288) | hypothetical protein MAV_1660 |
| M. thermoresistible (build 8) | TH_3271 | - | 1e-107 | 68.51% (289) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3372 | - | 6e-46 | 39.02% (287) | hypothetical protein MUL_3372 |
| M. vanbaalenii PYR-1 | Mvan_5745 | - | 1e-110 | 68.81% (295) | hypothetical protein Mvan_5745 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2748c|M.bovis_AF2122/97 ------MASVEF-ATILALG--AALLAGIGYVTLQRSARQVTAEEYVGHL
Rv2729c|M.tuberculosis_H37Rv ------MASVEF-ATILALG--AALLAGIGYVTLQRSARQVTAEEYVGHF
MMAR_1984|M.marinum_M ------MTTTEI-AAVLALG--AAFASALGNVTRQRSAQEITDEQ-VGHL
MUL_3372|M.ulcerans_Agy99 ------MTTTEI-AAVLALG--AAFASALGNVTRQRSAQEITDEQ-VGHL
MAV_1660|M.avium_104 ------MPRTEIIAALLALS--SALCVATGDVLQQRAAHCITDRE-LGPV
MSMEG_6539|M.smegmatis_MC2_155 ------MLGNSI-AVLLALC--AAIFMAVGIVVRQRATLDVPAGKGVSTV
TH_3271|M.thermoresistible__bu ------VLGQGL-AVLLAVC--AAVFAAVGIVVRQRATMDVPPEKGVSTV
Mflv_1077|M.gilvum_PYR-GCK ----------------------------MGIVVRQRATLDVPAEDGISAV
Mvan_5745|M.vanbaalenii_PYR-1 MCCPCRVIGHGV-TVLLAFL--AAVFLAVGIVVRQRATLDVPAEHGVSPV
MAB_2553|M.abscessus_ATCC_1997 -------MLIGIAAAMLACFGYGTASVLQGYGARQSSTEVGVAADHGPSL
* * :: . .
Mb2748c|M.bovis_AF2122/97 TLFHLSLRHALWWLGSLAAVASFTLQAIALTMGSVVLVQSLQATALLFAL
Rv2729c|M.tuberculosis_H37Rv TLFHLSLRHALWWLGSLAAVASFTLQAIALTMGSVVLVQSLQATALLFAL
MMAR_1984|M.marinum_M ELFWLSLRDSRWWLGALGAIANYLLQAAALTLGSVVLVSALQVTALLFAL
MUL_3372|M.ulcerans_Agy99 ELFWLSLRDSRWWLGALGAIANYLLQAAALTLGSVVLVSALQVTALLFAL
MAV_1660|M.avium_104 ELFAGLLRSQRWWWGAVLLLASIGLQAAALGGGSVLLVQALLMFSVLFAL
MSMEG_6539|M.smegmatis_MC2_155 MLRTLLRR-KLWWAGTISAVMGYGFQALALGFGSLLLVQPVLVSALLFAL
TH_3271|M.thermoresistible__bu MIATLVRR-RLWWIGTGAAVAGYALQAAALAFGSLILVQPILVSALLFAL
Mflv_1077|M.gilvum_PYR-GCK MFQTLIRR-PLWWAGTAAAIAGFVFQALALANGSLLLVQPILVSALLFAL
Mvan_5745|M.vanbaalenii_PYR-1 MIQTLVRR-PLWWAGTASAVAGFVFQALALANGSLLLVQPILVSALLFAL
MAB_2553|M.abscessus_ATCC_1997 KSTVAAALTPAFIAGMVLDLVGFAGSLVSARLIPLFLSQTIMSANLAVTA
: * : . . : .:.* ..: : .:
Mb2748c|M.bovis_AF2122/97 LIDARLTHHRCTPREWMWAVLLAGAVAVIVMSGNPAAGTTRAPFSTWAVV
Rv2729c|M.tuberculosis_H37Rv LIDARLTHHRCTPREWMWAVLLAGAVAVIVMSGNPAAGTTRAPFSTWAVV
MMAR_1984|M.marinum_M PIYARVTRRRVTRWEWTWAVLLAVSLAVVVIVGDPDVGVSRGSLATWMVV
MUL_3372|M.ulcerans_Agy99 PIYARVTRRRVTRWEWTWAVLLAVSLAVVVIVGDPDVGVSRGSLATWMVV
MAV_1660|M.avium_104 PINARLSHRAVTAGEWVWAALLTAAVTVVVVVGNPQTGHAGAPLRTWAVV
MSMEG_6539|M.smegmatis_MC2_155 PLSARLARRTVSRADWMWALLLTGALTVFVLLARTSTGTYSVSVSTTVTV
TH_3271|M.thermoresistible__bu PLSARLGARRVSRGEWAWAAVLTVALAAFVLIAKPKPGDYQASIPGTVVV
Mflv_1077|M.gilvum_PYR-GCK PLSARLAHRKVSRGEWAWAVLLTVALAVFVVLAKASPGDYEASLTTSIMV
Mvan_5745|M.vanbaalenii_PYR-1 PLSARLAHRRVTRAEWAWAVLLTLALAVFVVLAKASPGDYEASLSTSAVV
MAB_2553|M.abscessus_ATCC_1997 VLGIAVLGVRLQRRDWLAICAVLLSLCVLAATAGPRGEDSAPPAIHWGVL
: : :* : :: ... . . . :
Mb2748c|M.bovis_AF2122/97 AVVVVPAVVLCVVGARIASGSLSAVLLAVASSATLAVFTVLTKGVVTELG
Rv2729c|M.tuberculosis_H37Rv AVVVVPAVVLCVVGARIASGSLSAVLLAVASSATLAVFTVLTKGVVTELG
MMAR_1984|M.marinum_M ASVLGPVMVFCVLGARIWSGSVAAVLLAVVSGCSLALFAVLTKAVVEVVG
MUL_3372|M.ulcerans_Agy99 ASVLGPVMVFCVLGARIWSGSVAAVLLAVVSGCSLALFAVLTKAVVEVVG
MAV_1660|M.avium_104 ALVLGPLLAACVVAGRVRGGAVAAVLFAFVSGSLWGVFAVLTKEVMARLG
MSMEG_6539|M.smegmatis_MC2_155 AVVCTSVVALCVVLATRSSNWRRAVLLAAAVGVMFGVVAVLTKIVMHLVA
TH_3271|M.thermoresistible__bu TVACVAGVTVCVVLAVRSLGWRRALLLATAVGVLFGVVAVLTKVWLHKLF
Mflv_1077|M.gilvum_PYR-GCK AVVCTVAVLACVIVAARFVGWVRAVLLAVAVGVLFGVVAVLTKLVMHMLV
Mvan_5745|M.vanbaalenii_PYR-1 AVVCTVAVLACVIVATRIAGWIRAVLLAVAVGVLFGVVAVLTKLVMHVLT
MAB_2553|M.abscessus_ATCC_1997 TVSIS--VLLTGIALIRLLGTRAAIPAGLLAGILYGGMAVAVRVVGGVDP
: : : . *: . . . .:* .:
Mb2748c|M.bovis_AF2122/97 -EGFATLIRTPELYAWILVLPIGLMLQQSSLRVGALTASLPTITVARPVI
Rv2729c|M.tuberculosis_H37Rv -EGFATLIRTPALYAWILVLPIGLMLQQSSLRVGALTASLPTITVARPVI
MMAR_1984|M.marinum_M -GGAGALFRAPEFYVWIAAALGGMIFQQSSFRAGTLTASLPTVVVAKPLM
MUL_3372|M.ulcerans_Agy99 -GGAGALFRAPEFYVWIAAALGGMIFQQSSFRAGTLTASLPTVVVAKPLM
MAV_1660|M.avium_104 -QGVGVVTRTPELYACILVALGGVVWSQAAFRAGPLTASMPTLQVSQPVV
MSMEG_6539|M.smegmatis_MC2_155 DGDALATFTTPALYLVVVLGVLATLLQQSAFHAGSLQTSVPTMLVLEPVV
TH_3271|M.thermoresistible__bu TGGVAVGVTSPLLYLLILLGVLATLLQQSAFHAGSLQASVPTMLVLEPVV
Mflv_1077|M.gilvum_PYR-GCK HEGLVAVLTAPVLYLLALLGVVAMLIQQSAFHAGSLQTSVPTMLVLEPVI
Mvan_5745|M.vanbaalenii_PYR-1 NEGFVAVLTTPVLYLLALLGVVATLLQQSAFHAGSLQTSVPTMLVLEPMI
MAB_2553|M.abscessus_ATCC_1997 -LSLTVLLADPAAYGVVIAGVGGFYLFTVALQIGSVNGAAAALVVGETVV
. . * * . ::: *.: : .:: * ..::
Mb2748c|M.bovis_AF2122/97 ASVLGITVLDEVLHTGRVALVALVAVVVVVVVATVALAR---DEVAMMTV
Rv2729c|M.tuberculosis_H37Rv ASVLGITVLDEVLHTGRVALVALVAAVVVVVVATVALAR---DEVAMMTV
MMAR_1984|M.marinum_M GAVLGMVVLGETLDTTGPGLVVVAFGVVLVLVATIALARGEAEKVAEEAV
MUL_3372|M.ulcerans_Agy99 GAVLGMVVLGETLDTTGPGLVVVAFGVVLVLVATIALARGEAEKVAEEAV
MAV_1660|M.avium_104 ASVLGVVVLGETLDTGRAGTVALLVAVVVMTAAIIKLAR-------VEAV
MSMEG_6539|M.smegmatis_MC2_155 AVLLGSVVLGEHLSIAGWQPVALTLAIAAMIAATIALGRD--EGAYEETL
TH_3271|M.thermoresistible__bu AVALGAAVLGEHLDINGWEPVPLALAGAAMFAATVALGRN--EGAYEERL
Mflv_1077|M.gilvum_PYR-GCK AVVLGAVVLGEHLNAGHWDLVAIAVATIAMAAGTVALGRG--EGAYEEQL
Mvan_5745|M.vanbaalenii_PYR-1 AVILGAVVLGEHLNAGRWDAVALVVSTVAMVAGTIALGRD--EGAYEEQL
MAB_2553|M.abscessus_ATCC_1997 PGVIGVVLLGDSARPGMGWLVAVAFLGAVVSAVAVAVFG-----AAEHSR
:* .:*.: * : : . : :
Mb2748c|M.bovis_AF2122/97 SAGELGAAG-QLAVR---------------------
Rv2729c|M.tuberculosis_H37Rv SAGELGAAG-QLAVR---------------------
MMAR_1984|M.marinum_M ESGHIPATMPHRAVTMHFGPADDDVQTDRLRRAPAG
MUL_3372|M.ulcerans_Agy99 ESGHIPATMPHRAVTMHFGPADDDVQTDRLRRAPAG
MAV_1660|M.avium_104 ATRDRAEAQLHQPA----------------------
MSMEG_6539|M.smegmatis_MC2_155 EAEMEAKTVTG-------------------------
TH_3271|M.thermoresistible__bu EVAALAQQSRSQQVRT--------------------
Mflv_1077|M.gilvum_PYR-GCK EAD-QRTAQAASKSG---------------------
Mvan_5745|M.vanbaalenii_PYR-1 EADLAQKAQAASGNG---------------------
MAB_2553|M.abscessus_ATCC_1997 PVAPDPAR----------------------------