For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDHMPEPTLPDRTPAIDCDGLTHRYGQFTAVEDLTLQVRAGETLGLLGPNGAGKTTAVRVLTTLTPVQQ GRVRIFGLDTRAHTMDIRHNIGYVPQQLSIEAALTGRQNVELFARLYDVPRSERADRVAAALEAMQLLDV ADTVAGTYSGGMVRRLELAQALVNRPLLLILDEPTVGLDPIARDSVWTQVRAMQAEFGMTVLLTTHYMGE ADALCDRVALMHHGRLQAVGSPDELKRAVSSQASDDAGETTLEDVFRHYAGSDLEGGADPRRSGLGEVRS TRRTARRVS
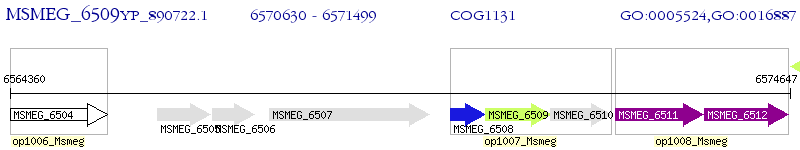
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6509 | - | - | 100% (289) | ABC-type drug export system, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_6766 | - | 1e-37 | 40.17% (229) | ABC transporter, ATP-binding protein, NodI family protein |
| M. smegmatis MC2 155 | MSMEG_3119 | - | 8e-36 | 40.91% (220) | ABC transporter, ATP-binding subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2961 | drrA | 5e-46 | 42.55% (235) | daunorubicin-DIM-transport ATP-binding protein ABC |
| M. gilvum PYR-GCK | Mflv_1936 | - | 1e-47 | 43.95% (223) | daunorubicin resistance ABC transporter ATPase subunit |
| M. tuberculosis H37Rv | Rv2936 | drrA | 6e-46 | 42.55% (235) | daunorubicin-DIM-transport ATP-binding protein ABC |
| M. leprae Br4923 | MLBr_02352 | - | 1e-44 | 41.70% (235) | putative antibiotic resistance efflux protein |
| M. abscessus ATCC 19977 | MAB_1858 | - | 3e-32 | 38.43% (229) | antibiotic ABC transporter ATP-binding protein |
| M. marinum M | MMAR_1771 | drrA | 2e-46 | 40.32% (248) | daunorubicin-DIM-transport ATP-binding protein ABC |
| M. avium 104 | MAV_5181 | - | 1e-105 | 72.10% (276) | daunorubicin resistance ABC transporter ATPase subunit |
| M. thermoresistible (build 8) | TH_2745 | - | 1e-31 | 35.56% (225) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_2014 | drrA | 1e-46 | 40.32% (248) | daunorubicin-DIM-transport ATP-binding protein ABC |
| M. vanbaalenii PYR-1 | Mvan_4796 | - | 2e-45 | 43.56% (225) | daunorubicin resistance ABC transporter ATPase subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6509|M.smegmatis_MC2_155 ------MTDHMPEPTLPDRTPAIDCDGLTHRYG---QFTAVEDLTLQVRA
MAV_5181|M.avium_104 ------MTGPPP--------MAIDAKHLTYRYG---QFTAVDDVTLQVRP
MMAR_1771|M.marinum_M ---------------MRNNDLAVEVRGVRKAFG---DVVALNDISFEVGR
MUL_2014|M.ulcerans_Agy99 ---------------MRNNDLAVEVRGVRKAFG---DVVALNDISFEVGR
Mb2961|M.bovis_AF2122/97 ---------------MRNDDMAVVVNGVRKTYGKG-KIVALDDVSFKVRR
Rv2936|M.tuberculosis_H37Rv ---------------MRNDDMAVVVNGVRKTYGKG-KIVALDDVSFKVRR
MLBr_02352|M.leprae_Br4923 ---------------MRNNDKAVMVRGVRKTYGKG-KIIALDDVSFDVDR
Mflv_1936|M.gilvum_PYR-GCK ----------------MPFPPAIEALDLVKRFG---DQTAVAGVSFVVPP
Mvan_4796|M.vanbaalenii_PYR-1 MADKSVGPMSVCAYTVDAMESAIEAIDLVKRFGDTLDNPAVDGVSFAVPP
MAB_1858|M.abscessus_ATCC_1997 ------------------MTEVIQVRELTFTYPKT-PEPAVRGMDFTVGE
TH_2745|M.thermoresistible__bu ---------MTTTSPRFDAEPAVEIHGLKKSFG---RTRAVDGLDLRVER
.: : : *: .: : *
MSMEG_6509|M.smegmatis_MC2_155 GETLGLLGPNGAGKTTAVRVLTTLTPVQQGRVRIFGLDTRAHTMDIRHNI
MAV_5181|M.avium_104 GETMGLLGPNGAGKTTMVRMLTTLAPVQQGELHIFGLDARRRTTDIRANI
MMAR_1771|M.marinum_M GEVIGLLGPNGAGKTTMVDILSTLTRPDAGSAHVAGSDVVAQPAGVRRSI
MUL_2014|M.ulcerans_Agy99 GEVIGLLGPNGAGKTTMVDILSTLTRPDAGSAHVAGYDVVAQPAGVRRSI
Mb2961|M.bovis_AF2122/97 GEVIGLLGPNGAGKTTMVDILSTLTRPDAGSAIIAGYDVVSEPAGVRRSI
Rv2936|M.tuberculosis_H37Rv GEVIGLLGPNGAGKTTMVDILSTLTRPDAGSAIIAGYDVVSEPAGVRRSI
MLBr_02352|M.leprae_Br4923 GEVIGLLGPNGAGKTTMVDILSTLTRPDRGLATVAGYDVVSEPAGVRRSI
Mflv_1936|M.gilvum_PYR-GCK GTVLGLLGPNGAGKTTTVRMMTTLTEPTSGTARVAGFDVRTHPDEVRRHM
Mvan_4796|M.vanbaalenii_PYR-1 GTVLGLLGPNGAGKTTTVRMMTTLTEPTSGTARVAGYDVRAQPDDVRRHM
MAB_1858|M.abscessus_ATCC_1997 GEIFGFLGPSGAGKSTTQKLLIGLLRGHGGRATVWGREPGAWGRNYYQRI
TH_2745|M.thermoresistible__bu GSVAGFLGPNGSGKSTTIRVLLGLLRADGGTARLLGGDPWHDAVELHRRL
* *:***.*:**:* :: * * : * : :
MSMEG_6509|M.smegmatis_MC2_155 GYVPQQLSIEAALTGRQNVELFARLYDVPRSERADRVAAALEAMQLLDVA
MAV_5181|M.avium_104 GYVPQQLSVEPALTGRQNVAWFARLYGVPRAERAERVDQALAAMDLLDVA
MMAR_1771|M.marinum_M MVTGQQVAVDDGLTGEQNLVLFGRLYGLRKSRARQRAQELLDQFGLTYAG
MUL_2014|M.ulcerans_Agy99 MVTGQQVAVDDGLTGEQNLVLFGRLYGLRKSRARQRAQELLDQFGLTYAG
Mb2961|M.bovis_AF2122/97 MVTGQQVAVDDALSGEQNLVLFGRLWGLSKSAARKRAAELLEQFSLVHAG
Rv2936|M.tuberculosis_H37Rv MVTGQQVAVDDALSGEQNLVLFGRLWGLSKSAARKRAAELLEQFSLVHAG
MLBr_02352|M.leprae_Br4923 MVTGQQVAVDDQLSGEQNLILFGRLYGLSKSVARVRTQELLEQFDLLHAG
Mflv_1936|M.gilvum_PYR-GCK GLTGQVATVDELLTGRENIRMIGGLYGIRRRDLKNLGDQLLHQFSLTDAA
Mvan_4796|M.vanbaalenii_PYR-1 GLTGQVATVDELLTGRENIRMIGGLYGIRRKDLDRLGDQLLHQFSLTDAA
MAB_1858|M.abscessus_ATCC_1997 GVSFELPNHYQKLTALENLRFFASLYAGPTRDP----MELLDAVGLADDA
TH_2745|M.thermoresistible__bu AYVPGDVTLWPNLTGGQAIDFLAGLRDNFDTKRR---DELIERFELD--P
*:. : : :. * : . *
MSMEG_6509|M.smegmatis_MC2_155 DTVAGTYSGGMVRRLELAQALVNRPLLLILDEPTVGLDPIARDSVWTQVR
MAV_5181|M.avium_104 DRLASAYSGGMIRRLEVAQALVNRPSLLVLDEPTVGLDPIARDGVWEHVQ
MMAR_1771|M.marinum_M KRLVRTYSGGMRRRIDIACGLVVEPQVAFLDEPTTGLDPRSRQAIWDLVT
MUL_2014|M.ulcerans_Agy99 KRLVRTYSGGMRRRIDIACGLVVEPQVAFLDEPTTGLDPRSRQAIWDLVT
Mb2961|M.bovis_AF2122/97 KRRVGTYSGGMRRRIDIACGLVVQPQVAFLDEPTTGLDPRSRQAIWDLVA
Rv2936|M.tuberculosis_H37Rv KRRVGTYSGGMRRRIDIACGLVVQPQVAFLDEPTTGLDPRSRQAIWDLVA
MLBr_02352|M.leprae_Br4923 KRRVGTYSGGMRRRIDIACGLVVQPQVAFLDEPTTGLDPRSRQAIWDLVA
Mflv_1936|M.gilvum_PYR-GCK DRPVRSYSGGMRRRLDLAVSLLASPPVLFLDEPTTGLDPRSRSELWDVLR
Mvan_4796|M.vanbaalenii_PYR-1 DRVVRSYSGGMRRRLDLAVSLLASPPVLFLDEPTTGLDPRSRSELWDVLR
MAB_1858|M.abscessus_ATCC_1997 NTRVGAFSKGMQMRLTFVRALINDPELLFLDEPTSGLDPVNARKVKDMVL
TH_2745|M.thermoresistible__bu DKKARTYSKGNRQKVALVAAFSTNAEIYILDEPTSGLDPLMDKAFQMCVR
. . ::* * :: .. .: . : .***** **** . :
MSMEG_6509|M.smegmatis_MC2_155 AMQAEFGMTVLLTTHYMGEADALCDRVALMHHGRLQAVGSPDELKRAVSS
MAV_5181|M.avium_104 KMQARFAMTVLLTTHYMEEADALCDRVALMHRGALRAVGAPDELKARIS-
MMAR_1771|M.marinum_M SFKK-LGIATLLTTQYLEEADALSDRIILIDHGKIIADGTANELKHRAGD
MUL_2014|M.ulcerans_Agy99 SFKK-LGIATLLTTQYLEEADALSDRIILIDHGKIIADGTANELKHRAGD
Mb2961|M.bovis_AF2122/97 SFKK-LGIATLLTTQYLEEADALSDRIILIDHGIIIAEGTANELKHRAGD
Rv2936|M.tuberculosis_H37Rv SFKK-LGIATLLTTQYLEEADALSDRIILIDHGIIIAEGTANELKHRAGD
MLBr_02352|M.leprae_Br4923 SFKQ-IGIATLLTTQYLEEADELSDRIILIDHGTVIAEGTASELKHRVGE
Mflv_1936|M.gilvum_PYR-GCK GLVA-QGTTLLLTTQYLEEADQLADNIVVIDRGRIIAEGSPLHLKQQAGR
Mvan_4796|M.vanbaalenii_PYR-1 GLVA-QGTTLLLTTQYLEEADQLADNIVVIDRGHIIAEGSPLELKQQAGR
MAB_1858|M.abscessus_ATCC_1997 ELKA-RGRTIFITTHAMSAADELCDRVAFVVDGRIVAMNSPAELKLEYGR
TH_2745|M.thermoresistible__bu EAAD-NGAAVLLSSHILAEVERLCDSVTIIRSGRTVRAGRLDELRHLMRT
. : ::::: : .: *.* : .: * . .*:
MSMEG_6509|M.smegmatis_MC2_155 ---------------------------------------QASDDAGETTL
MAV_5181|M.avium_104 ---------------------------------------RVSQHA---TL
MMAR_1771|M.marinum_M TFCEIVPRHLKDLDATVAALGSLLPEHNRAMVTSESDRITMPAPDGTRTL
MUL_2014|M.ulcerans_Agy99 TFCEIVPRHLKDLDATVAALGSLLPEHNRAMVTSESDRITMPAPDGTRTL
Mb2961|M.bovis_AF2122/97 TFCEIVPRDLKDLDAIVAALGSLLPEHHRAMLTPDSDRITMPAPDGIRML
Rv2936|M.tuberculosis_H37Rv TFCEIVPRDLKDLDAIVAALGSLLPEHHRAMLTPDSDRITMPAPDGIRML
MLBr_02352|M.leprae_Br4923 TFCEIVPRDLKDLGTVVAALGSLLPERSRAMLTPTSDRITMPAPDGTQTL
Mflv_1936|M.gilvum_PYR-GCK ASLVVTVADAADLESARGLLA---RTGAEVYVDAGARRLTATA-DGLDDM
Mvan_4796|M.vanbaalenii_PYR-1 ASLVVTVGDAADLEGARTLLG---QTGAEVFVDAGARRLTASA-DGLDDM
MAB_1858|M.abscessus_ATCC_1997 RSVRVEYRG-------------------------AAGELAGQDFPLDGLA
TH_2745|M.thermoresistible__bu TVKVRTRAPIDALRTAP--------------YIHEFSTDDGLHTFSVDRT
MSMEG_6509|M.smegmatis_MC2_155 EDVFRHYAGSDLEG--GADPRRSG-LGEVRSTRRTARR-------VS---
MAV_5181|M.avium_104 EDVFRHYAASDLGGDDAAHPRPSGSIRQVRSSRKVARR-------VG---
MMAR_1771|M.marinum_M IEAARRIDEANIELADIALRRPSLDDVFLSMTTDPSEAHALSHVASGAML
MUL_2014|M.ulcerans_Agy99 IEAARKIDEANIELADIALRRPSLDDVFLSMTTDPSEAHVLSHVASGAML
Mb2961|M.bovis_AF2122/97 VEAARRIDEARIELADIALRRPSLDDVFLAMTTDPTES--LTHLVSGSAR
Rv2936|M.tuberculosis_H37Rv VEAARRIDEARIELADIALRRPSLDHVFLAMTTDPTES--LTHLVSGSAR
MLBr_02352|M.leprae_Br4923 IEAARRIDEVSVELADIALRRPSLDDVFLSMTTDPSES--LSHLAAGAMR
Mflv_1936|M.gilvum_PYR-GCK VKVAGWLRDSGITVDDIGLSRPSLDDVFLMLTGHRTDD----TEEVGA--
Mvan_4796|M.vanbaalenii_PYR-1 IRVAEWLRDSGIAVDDIGLSRPSLDDVFLTLTGHRTED----SEEIGA--
MAB_1858|M.abscessus_ATCC_1997 DNDAFHAVLRGHHVETIHSEEASLDDVFVAVTGRRLA-------------
TH_2745|M.thermoresistible__bu DLDRTMTELTTLGIEELTVTPASLEDLFLREYQEVPR-------------
* :