For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAAVEVRGLSKSYGRRPAVRDLDLDVGYGEIFAILGPNGAGKTTTVEILEGNRRRDSGEVLVLGEDPGRA GRQWRARIGIVLQEVSDAGMLTVGETVQMFSACYGTARAPADVLELVGLRHASRSRVRTLSGGQRRRLDV ALGIIGGPELVFLDEPTTGFDPEARQQFWELIRLLAADGTTIVLTTHYLEEAAALADRVAVIADGNVVAV DSPTRLTRHVDTAATVRWMEGGHVHEERTEFPTELIRQRSRVGAELAQLTVTRPSLEDAYLHLIGRT*
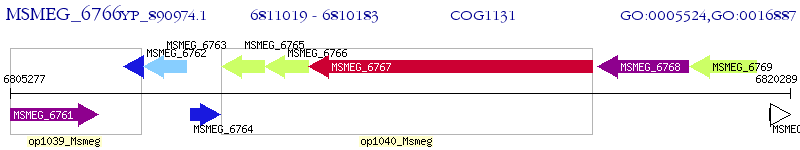
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6766 | - | - | 100% (278) | ABC transporter, ATP-binding protein, NodI family protein |
| M. smegmatis MC2 155 | MSMEG_3119 | - | 8e-49 | 38.54% (301) | ABC transporter, ATP-binding subunit |
| M. smegmatis MC2 155 | MSMEG_1502 | - | 1e-39 | 41.55% (219) | ABC transporter, ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1493c | - | 9e-49 | 47.44% (215) | unidentified antibiotic-transport ATP-binding protein ABC |
| M. gilvum PYR-GCK | Mflv_3680 | - | 1e-49 | 40.53% (301) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1458c | - | 9e-49 | 47.44% (215) | unidentified antibiotic-transport ATP-binding protein ABC |
| M. leprae Br4923 | MLBr_00590 | - | 5e-49 | 47.42% (213) | putative ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_2752 | - | 6e-43 | 38.21% (280) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_2263 | - | 5e-48 | 47.44% (215) | antibiotic ABC transporter ATP-binding protein |
| M. avium 104 | MAV_3321 | - | 8e-51 | 49.77% (215) | ABC transporter, ATP-binding subunit |
| M. thermoresistible (build 8) | TH_3667 | - | 8e-31 | 36.60% (235) | PUTATIVE ferric cations import ATP-binding protein FbpC |
| M. ulcerans Agy99 | MUL_1859 | - | 3e-48 | 47.44% (215) | antibiotic ABC transporter ATP-binding protein |
| M. vanbaalenii PYR-1 | Mvan_2730 | - | 2e-51 | 49.54% (218) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1493c|M.bovis_AF2122/97 ---------------------MN---RAPDT---PEVVLRLRGVCKRYGS
Rv1458c|M.tuberculosis_H37Rv ---------------------MN---RAPDT---PEVVLRLRGVCKRYGS
MLBr_00590|M.leprae_Br4923 ---------------------MSSACTVPET---PEVVIRLRGVCKHYGS
MAV_3321|M.avium_104 --------------------------------------MRLRGVSKHYGS
MMAR_2263|M.marinum_M ---------------------MSSDPKYPAAGRGTEIVVRLRGVGKQYGS
MUL_1859|M.ulcerans_Agy99 -------------------------------------MVRLRGVGKQYGS
MAB_2752|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3680|M.gilvum_PYR-GCK ---------------------MTSVSPGAVPDGSPSAPVVLRGVSKRYGS
Mvan_2730|M.vanbaalenii_PYR-1 ---------------------MS----------SQSAPVQVRGVSKRYGA
MSMEG_6766|M.smegmatis_MC2_155 ----------------------------------MSAAVEVRGLSKSYGR
TH_3667|M.thermoresistible__bu VIKTMKTKPADTSGAGPAVSNPAVSNPDVSAPQLRGADVEVQGLHKSFGA
Mb1493c|M.bovis_AF2122/97 ITAVSNLDLDVHDAEVMALLGPNGAGKTTTVEMCEGFVRPDAGSIEVLGL
Rv1458c|M.tuberculosis_H37Rv ITAVSNLDLDVHDAEVMALLGPNGAGKTTTVEMCEGFVRPDAGSIEVLGL
MLBr_00590|M.leprae_Br4923 ILAVSNLDLDIHSAEVLALLGPNGAGKTTTVEMCEGFVRPDAGTIEVLGL
MAV_3321|M.avium_104 TTAVRDLDLEVHAAEVLALLGPNGAGKTTTVEMCEGFVRPDAGTLEVLGL
MMAR_2263|M.marinum_M VTAVSNLDLEVRAAEVMALLGPNGAGKTTTVEMCEGFVRPDAGSIEVLGL
MUL_1859|M.ulcerans_Agy99 VTAVSNLDLEVRAAEVMALLGPNGAGKTTTVEMCEGFVRPDAGSIEVLGL
MAB_2752|M.abscessus_ATCC_1997 ---MDSLDLEVHAAQVFALLGPNGAGKTTTVELCEGFLHPDEGTVEVLGM
Mflv_3680|M.gilvum_PYR-GCK TTAVAGLDLVVERAEVFALLGPNGAGKTTTVEMCEGFIKPDAGSLRILGL
Mvan_2730|M.vanbaalenii_PYR-1 TTAVASLDLEVQTAEVFALLGPNGAGKTTTVEMCEGFMKPDSGSIRILGL
MSMEG_6766|M.smegmatis_MC2_155 RPAVRDLDLDVGYGEIFAILGPNGAGKTTTVEILEGNRRRDSGEVLVLGE
TH_3667|M.thermoresistible__bu KAVLRGVDLTVSAGTLTAVLGPSGCGKTTLLRILAGFDDPDAGTVRIGGH
: .:** : . : *:***.*.**** :.: * * * : : *
Mb1493c|M.bovis_AF2122/97 DPI---TDNARLRARIGVMLQGGGGYPAARAGEMLDL---VASYAANPLD
Rv1458c|M.tuberculosis_H37Rv DPI---TDNARLRARIGVMLQGGGGYPAARAGEMLDL---VASYAANPLD
MLBr_00590|M.leprae_Br4923 DPI---TDNARLRARIGVMLQGGGGYPAARAGEMLNL---VASYAADPLD
MAV_3321|M.avium_104 DPI---ADNARLRPRIGVMLQGGGGYPAARAGEMLNL---VAAYAADPLD
MMAR_2263|M.marinum_M DPI---ADNARLRPRIGVMLQGGGGYPAARAGEMLDL---VASYAADPLD
MUL_1859|M.ulcerans_Agy99 DPI---ADNARLRPRIGVMLQGGGGYPAARAGEMLDL---VASYAADPLD
MAB_2752|M.abscessus_ATCC_1997 DPV---AKNSAVRSRIGVMLQGGGAYPTARASEMLGL---VASYSANPLD
Mflv_3680|M.gilvum_PYR-GCK DPI---VDNAEVRARTGVMLQGGGAYPAARAGEMLDL---VAAYAADPLD
Mvan_2730|M.vanbaalenii_PYR-1 DPI---ADNAEVRARTGVMLQGGGAYPAARAGEMLEL---VAAYAADPLD
MSMEG_6766|M.smegmatis_MC2_155 DPG---RAGRQWRARIGIVLQEVSDAGMLTVGETVQM---FSACYGTARA
TH_3667|M.thermoresistible__bu TLAGGGAPVPAHRRRVGLMPQEGALFPHLSVAENIAFGLRRTRRAGRADT
* * *:: * . ..* : : : . .
Mb1493c|M.bovis_AF2122/97 PHWLLDTLGLTEAARTTYRRLSGGQQQRLALACALVGRPQLVFLDEPTAG
Rv1458c|M.tuberculosis_H37Rv PHWLLDTLGLTEAARTTYRRLSGGQQQRLALACALVGRPQLVFLDEPTAG
MLBr_00590|M.leprae_Br4923 PEWLLETLGLTDVARTTYRRLSGGQQQRLALACTLVGRPELVFLDEPTAG
MAV_3321|M.avium_104 PAWLLDTLGLTDAARTTYRRLSGGQQQRLALACALVGRPELVFLDEPTAG
MMAR_2263|M.marinum_M PRWLLDTLGLTDAARTTYRRLSGGQQQRLALACALVGRPELVFLDEPTAG
MUL_1859|M.ulcerans_Agy99 PRWLLDTLGLTDAARTTYRRLSGGQQQRLALACALVGRPELVFLDEPTAG
MAB_2752|M.abscessus_ATCC_1997 PQWLLDTLGLTDAARTPYRRLSGGQQQRLALACALVGRPELVFLDEPTAG
Mflv_3680|M.gilvum_PYR-GCK PTWLLDTLGLTDAARTTYRRLSGGQQQRLALACAVVGRPELVFLDEPTAG
Mvan_2730|M.vanbaalenii_PYR-1 PQWLLDTLGLTDAARTTYRRLSGGQQQRLALACAVIGRPELVFLDEPTAG
MSMEG_6766|M.smegmatis_MC2_155 PADVLELVGLRHASRSRVRTLSGGQRRRLDVALGIIGGPELVFLDEPTTG
TH_3667|M.thermoresistible__bu VRYWLEVVGLEGAAEARPHELSGGQQQRVALARALAAEPRVLLLDEPFVA
*: :** .:.: : *****::*: :* : . *.:::**** ..
Mb1493c|M.bovis_AF2122/97 MDAHARVLVWELIDALR-RDGVTVVLTTHHLKEAEELADRLVIIDHGVTV
Rv1458c|M.tuberculosis_H37Rv MDAHARVLVWELIDALR-RDGVTVVLTTHHLKEAEELADRLVIIDHGVTV
MLBr_00590|M.leprae_Br4923 MDTHARLLVWELIDTLR-RDGVTVVLTTHQLKEAEELADRLVIIDHGRMI
MAV_3321|M.avium_104 MDAHARLLVWELIDALR-RDGVTVVLTTHQLKEAEELADRLVIIDHGATV
MMAR_2263|M.marinum_M MDAHARLLVWELIDALR-RDGVTVVLTTHQLKEAEELADRLVIIDHGVTV
MUL_1859|M.ulcerans_Agy99 MDAHARLLVWELIDALR-RDGVTVVLTTHQLKEAEELADRLVIIDHGVTV
MAB_2752|M.abscessus_ATCC_1997 MDAHARLVVWELIDCLR-RDGVSVVLTTHHMKEAEELADQLMIIDHGSVV
Mflv_3680|M.gilvum_PYR-GCK MDAHARIVVWELIDALR-RDGVTVVLTTHQLSEAEELADRILIIDNGAAV
Mvan_2730|M.vanbaalenii_PYR-1 MDAHARIVVWELIDALR-RDGVTVVLTTHQLAEAEELADRILIIDHGAAV
MSMEG_6766|M.smegmatis_MC2_155 FDPEARQQFWELIRLLA-ADGTTIVLTTHYLEEAAALADRVAVIADGNVV
TH_3667|M.thermoresistible__bu LDAGLRVRVREEIAAILRAAGTTAVLVTHDQSEALSLADSVALLMDGRLA
:*. * . * * : *.: **.** ** *** : :: .*
Mb1493c|M.bovis_AF2122/97 AAGTPAELMRSGAKDQLRFTAPPRLDLS---------------LLASALP
Rv1458c|M.tuberculosis_H37Rv AAGTPAELMRSGAKDQLRFTAPPRLDLS---------------LLASALP
MLBr_00590|M.leprae_Br4923 AAGTPTELMHTGAKDQLRFSAPPRLDLS---------------LLAFALP
MAV_3321|M.avium_104 AAGTPAELMRSGAKDQLRFSAPPRLDLS---------------LLSAALP
MMAR_2263|M.marinum_M AEGTPSELMRSGAKDQLRFTAPPRLDLS---------------LLVSALP
MUL_1859|M.ulcerans_Agy99 AEGTPSELMRSGAKDQLRFTAPPRLDLS---------------LLVSALP
MAB_2752|M.abscessus_ATCC_1997 AAGTPSELMQSGAENQLRFAAPPRLDLT---------------LLTSALP
Mflv_3680|M.gilvum_PYR-GCK ATGTPAELMRNGAENQLRFRAPKMLDLS---------------LLVAALP
Mvan_2730|M.vanbaalenii_PYR-1 ATGTPAELMRSGAENQLRFRAPRMLDLS---------------LLVAALP
MSMEG_6766|M.smegmatis_MC2_155 AVDSPTRLTR-------------HVDTA---------------ATVRWME
TH_3667|M.thermoresistible__bu QVGAPADIYERPATPAAARFVGATVELPGQARDGVVHTALGSHPARLPVP
.:*: : . :: . :
Mb1493c|M.bovis_AF2122/97 EGYQATELTPGEYLVEGPVDPQVLATVTAWCAQIDVLATDMRVEQRSLED
Rv1458c|M.tuberculosis_H37Rv EGYQATELTPGEYLVEGPVDPQVLATVTAWCAQIDVLATDMRVEQRSLED
MLBr_00590|M.leprae_Br4923 EDYKTTELTPGEYLVAGPIDPQVLATVTAWCAQINVLTTNMRVEQHSLED
MAV_3321|M.avium_104 EDYKATELTPGEYLVEGPVDPQVLATVTAWCARIDVLATDMRVEQRSLED
MMAR_2263|M.marinum_M EDYRVTEVTPGEYLVEGSVDPQVLAAVTAWCARIDVLATDMRVEQRSLED
MUL_1859|M.ulcerans_Agy99 EDYRVTEVTPGEYLVEGSVDPQVLAAVTAWCARIDVLATDMRVEQRSLED
MAB_2752|M.abscessus_ATCC_1997 EGYKATEVTPGEYRVEGVVDPQVLATVTAWCARMDVLTTDLRVEKRSLED
Mflv_3680|M.gilvum_PYR-GCK EGYRATEPTAGEYLVEGHIDPQVLATVTAWCARLDVLATDMRVEQRSLED
Mvan_2730|M.vanbaalenii_PYR-1 EAYRATETAPGEYLVEGHIDPQVLATVTAWCARLNVLATDMRVEQRSLED
MSMEG_6766|M.smegmatis_MC2_155 GGHVHEERT------------EFPTELIRQRSRVGAELAQLTVTRPSLED
TH_3667|M.thermoresistible__bu DGPVVVVLRPEELRLERSRDGVPAAAVQSVRFYGPQSAVQVRLADDTVVT
: : .:: : ::
Mb1493c|M.bovis_AF2122/97 VFLD-----------------------LTGRKLRQ-
Rv1458c|M.tuberculosis_H37Rv VFLD-----------------------LTGRKLRQ-
MLBr_00590|M.leprae_Br4923 VFLD-----------------------LTGRKLR--
MAV_3321|M.avium_104 VFLD-----------------------LTGRKLRQ-
MMAR_2263|M.marinum_M VFLD-----------------------LTGRKLRS-
MUL_1859|M.ulcerans_Agy99 VFLD-----------------------LTGRKLRS-
MAB_2752|M.abscessus_ATCC_1997 VFLD-----------------------LTGRELR--
Mflv_3680|M.gilvum_PYR-GCK VFLD-----------------------LTGRELRK-
Mvan_2730|M.vanbaalenii_PYR-1 VFLD-----------------------LTGRELRK-
MSMEG_6766|M.smegmatis_MC2_155 AYLH-----------------------LIGRT----
TH_3667|M.thermoresistible__bu VLTAGDAGVSVGEPVGVRVDGSVLAYPAPGRPPARS
. **