For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ITAPLAGGVIDVHAHWLPRELFDLPPGAPYGPLHDRDGQLHLGDLPLSIATDAMSDEAAIIDDMDRAGVG VRVLSAPPFAFPLRGGDDADRYAAEFNQALAKLVARGDGRLAGLGIVSLDAGPDFASGSVPRQLDHLAAV DGIAGVAIPPVVDGRSFDGAALRHVLTETARRDLAVLVHPMQIGRAEWSQFYLANLIGNPVETATAVATL ILSGLQEELPDLRICFLHGGGCAPGLLGRWSHGWTARADVRSRTSRPPAEAFRRLYFDTITHGVPQLELL TELAGEDRIVCGSDYPFDMAEVDPARFAVEHGPGQDSLIRAARAFLGIREARAGDTINPYRNPSATPVLA PDAT*
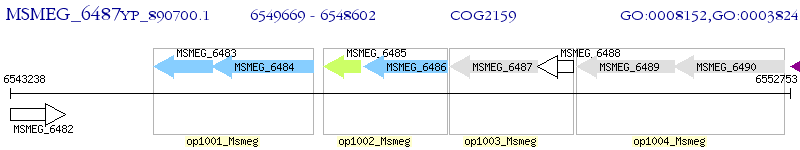
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6487 | - | - | 100% (355) | 2-hydroxy-3-carboxy-6-oxo-7-methylocta-2, 4-dienoate decarboxylase |
| M. smegmatis MC2 155 | MSMEG_6813 | - | 2e-09 | 26.67% (255) | amidohydrolase 2 |
| M. smegmatis MC2 155 | MSMEG_4063 | - | 1e-05 | 23.62% (254) | amidohydrolase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02013 | - | 3e-06 | 28.03% (132) | hypothetical protein MLBr_02013 |
| M. abscessus ATCC 19977 | MAB_0299 | - | 7e-21 | 31.65% (297) | putative amidohydrolase (aminocarboxymuconate-semialdehyde |
| M. marinum M | MMAR_0083 | - | 9e-11 | 27.61% (297) | amidohydrolase |
| M. avium 104 | MAV_0481 | - | 1e-06 | 23.29% (249) | amidohydrolase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2437 | - | 3e-10 | 29.67% (246) | hypothetical protein MUL_2437 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6487|M.smegmatis_MC2_155 MITAPLAGGVIDVHAHWLPRELFDLPPGAPYGPLHDRDGQLHLGDLPLSI
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
MAB_0299|M.abscessus_ATCC_1997 -----MTSVRIDIHAHLWSDEYLSLLDGYGR-PATDVHRGLGAG------
MAV_0481|M.avium_104 ----MRIITVEEHFQHPEVSARVAELTGPAVEGLAEFGNAFSLD------
MMAR_0083|M.marinum_M -------MTFVDVHFHLAPASFIRQKP----NRVFDIYPQLG--------
MUL_2437|M.ulcerans_Agy99 --MGTRLEPHRGRRIHLGADDLPGNQHLGPYSRGTAHHRQVGPVQDGGAF
MSMEG_6487|M.smegmatis_MC2_155 ATDAMSDEAAIID-------------------------------------
MLBr_02013|M.leprae_Br4923 ----------MLR-------------------------------------
MAB_0299|M.abscessus_ATCC_1997 --ATRDDLDGRFE-------------------------------------
MAV_0481|M.avium_104 PDSTARLGGNRLA-------------------------------------
MMAR_0083|M.marinum_M --------DGALG-------------------------------------
MUL_2437|M.ulcerans_Agy99 VAAARGQRGAALRPGKEFAIWRIDTHHHMIPPDYRRALRNAGVDGAGGRA
:
MSMEG_6487|M.smegmatis_MC2_155 -----------DMDRAGVGVRVLSAPP--FAFPLRGGDDADRYAAEFNQA
MLBr_02013|M.leprae_Br4923 -----------DVDREHIDMMVLYPSLGFCILRLDDPDFATRLARFYNQW
MAB_0299|M.abscessus_ATCC_1997 -----------LMDEAGVDLQILTATP--ASPHFEDQAAAVASARFINDE
MAV_0481|M.avium_104 -----------HMDQVGIDVQVVSHGN--GSPGTLQHPEAVELCRRVNND
MMAR_0083|M.marinum_M -----------RQELAAAGGRAFVSLP-WPVPPGADVEATATATSSCNDE
MUL_2437|M.ulcerans_Agy99 LPDWSPAGSLQAMAELTVGAAILSVSAP-ATAFLPQASDAAALARDINDY
. . : *:
MSMEG_6487|M.smegmatis_MC2_155 LAKLVARGDGRLAGLGIVSLDAGPDFASGSVPRQLDHLAAVDGIAGVAIP
MLBr_02013|M.leprae_Br4923 IGDYCAPTNGWLRGGGVTSMER------GQVAIDITNGVKELGIAVTLIP
MAB_0299|M.abscessus_ATCC_1997 YAKLSSEYPGRFGALASLPLPHVP-----AALEELARAIDELGMYGASIT
MAV_0481|M.avium_104 LAAQIAENPDRFRGFATLPLYDPA-----AAAEELRRCVGDLGFVGALIA
MMAR_0083|M.marinum_M LLAAAAEHVEYAGAMIAVDLTDPA-----RAVTEVRRTADAQAMAGVMIY
MUL_2437|M.ulcerans_Agy99 AADVVAGRPAQFGFFATVPMPHLD-----QSVAETVRALDDLKADGVVLL
: : : . . :
MSMEG_6487|M.smegmatis_MC2_155 PVVDGRSFDGAALRHVLTETARRDLAVLVHP----------MQIGRAE--
MLBr_02013|M.leprae_Br4923 PVLNASNLDHPYLGPFYAATVERGMAISIHA----------RYPFAAD--
MAB_0299|M.abscessus_ATCC_1997 TSVLGRSIADPIFDPLYAELNRRRAVLFVHP----------AGCGAESSL
MAV_0481|M.avium_104 GSYDGLFLDDERFDPILTAAEAVDFPIYVHPGLPEAPVSQRYYAGSWPAS
MMAR_0083|M.marinum_M VSPTSR-LDEEGLHEFYAELSARRLPLFLHP-----------ALEPPIGG
MUL_2437|M.ulcerans_Agy99 ANHAGVYLGEDGQDDLFTALNSRAAVVFVHP-------------AELPGP
. : . : : :*.
MSMEG_6487|M.smegmatis_MC2_155 WSQFYLANLIGNPVETATAVATLILSGLQEELPDLRICFLHGGGCAPGLL
MLBr_02013|M.leprae_Br4923 WC------------------------------------------------
MAB_0299|M.abscessus_ATCC_1997 ITDHSLTWSIGAPIEDTVAIMHLIVAGVPSRYPDMKIVTCHLGGALPMVL
MAV_0481|M.avium_104 VHMMFSGPAFGWHAEAGIHIVRLILSGALDRHPTLKLLSGHWGELAAFYL
MMAR_0083|M.marinum_M ATDWSLDASLSPPVVTSTAAARLMLSGRLDEHPGLTLIIPHLGGVLPYLA
MUL_2437|M.ulcerans_Agy99 AVPGVAPWAGDFLLDTTRAAHLLVRNGIRQRYPDINFVLSHAGGFVPYAS
MSMEG_6487|M.smegmatis_MC2_155 GRWSHGWTARADVRSRTSRPPAEAFRRLYFDTITHG-VPQLELLTELAGE
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
MAB_0299|M.abscessus_ATCC_1997 ERAHRQVTEWEATQCP--EAPRDAARRLWYDTVGHDHTPALRAAVASLGV
MAV_0481|M.avium_104 ERLDETLGLLPTGLER--APSDYYRQQVWITPSGMYNQNQLNFMTAELGA
MMAR_0083|M.marinum_M QRLIDQSGRGAALHDI----PTYLRTRTYVDTCSFH-PPALRCALDTIGP
MUL_2437|M.ulcerans_Agy99 HRLALVITADTGRSPAD---VLEEFSSFYFDTALSSSAAALPTLIAFAKP
MSMEG_6487|M.smegmatis_MC2_155 DRIVCGSDYPFDMAEVDPARFA------VEHGPGQDSLIRAARAFLGIRE
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
MAB_0299|M.abscessus_ATCC_1997 DRLIFGSDFPYQAGP------------------------RYVSSATYIEE
MAV_0481|M.avium_104 QRIIYSEDFPYVVRDN--------------------------VSTFLEQA
MMAR_0083|M.marinum_M SRLVLGSDYPFRGPVAR-------------------------AIADIADS
MUL_2437|M.ulcerans_Agy99 GHITFGSDWPFAPTAAGKLFAAGLETYPALDAAARGAIDRGSALSLFPRF
MSMEG_6487|M.smegmatis_MC2_155 ARAGDTINPYRNPSATPVLAPDAT-----------
MLBr_02013|M.leprae_Br4923 -----------------------------------
MAB_0299|M.abscessus_ATCC_1997 GLSAEDASAILDHNGAELLGLTGV-----------
MAV_0481|M.avium_104 GLSEADTHAIAHTNAEALLRI--------------
MMAR_0083|M.marinum_M GVDETFKESALIGNATAAIRTAAGAV---------
MUL_2437|M.ulcerans_Agy99 GVAAPPPQPSAAQRIRHTLSRTAMRGVARLMNSGG