For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLANLARLLPHKNDYTDVPRTWRQDVLAGITVGVVALPLALAFGISSGVGAAAGLITAVVAGLVAAVFGG SHVQVSGPTGAMAVVLAPIVAEHGLGSIALVTVLAGLLVLAAGITGLGRAVTFIPWPVIEGFTLGIAAII FLQQVPAALAQQAPSGERTLLAAFTVVRNADWGEAAKTLAVVGVVAVLMIVLPRVHSGIPASLTAVIVAT VLIVALDIPVATIGELPSHLPAPVLPHVDVNTLRTMFSAALAIAALAAIESLLSARVAATMSPTGPYDPN RELVGQGLASVASGAFGGMPATGAIARTAVNVRSGAATRLSAIVHSLVLLAVVYLATGPVSAIPLSALAG VLMVTSFRMISATTIRKIVRSTRSDALIFVLTAVITVCFDLIEAVEIGILVAAFFALRSVARRSSVVREE LPGPRRPGDDQIALLRLDGAMFFGAAERISNAITDAEHPHVSVVIIRMSQLGMLDATGAHTLAEITTELE SRGITVIIKGVRPEHMDLLANVGVFESLRHENHLLDSLDDAVAHARTHVARQPH
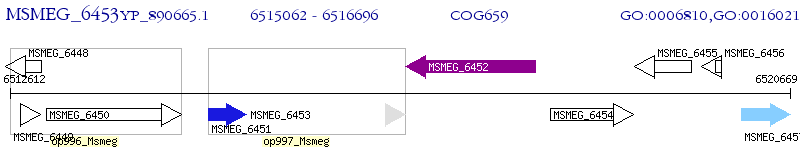
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6453 | - | - | 100% (544) | sulfate permease |
| M. smegmatis MC2 155 | MSMEG_1235 | - | 7e-59 | 31.80% (500) | sulfate permease |
| M. smegmatis MC2 155 | MSMEG_1448 | - | 8e-57 | 31.31% (495) | integral membrane transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1734 | - | 4e-63 | 33.47% (484) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1119 | - | 0.0 | 80.85% (543) | sulphate transporter |
| M. tuberculosis H37Rv | Rv1707 | - | 4e-63 | 33.47% (484) | transmembrane protein |
| M. leprae Br4923 | MLBr_02279 | - | 2e-52 | 31.91% (492) | putative transmembrane transport protein |
| M. abscessus ATCC 19977 | MAB_4448 | - | 2e-57 | 29.85% (526) | putative sulfate transporter |
| M. marinum M | MMAR_4750 | - | 2e-57 | 31.73% (498) | integral membrane transporter |
| M. avium 104 | MAV_0121 | - | 5e-52 | 34.55% (385) | carbonate dehydratase |
| M. thermoresistible (build 8) | TH_2104 | - | 0.0 | 78.48% (539) | sulfate permease |
| M. ulcerans Agy99 | MUL_0321 | - | 1e-55 | 31.53% (498) | integral membrane transporter |
| M. vanbaalenii PYR-1 | Mvan_5693 | - | 0.0 | 82.44% (541) | sulphate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1119|M.gilvum_PYR-GCK ------MITIGR--LGRLLPQRSDYTDLRRSWRADLLAGVTVGVVALPLA
Mvan_5693|M.vanbaalenii_PYR-1 ------MIAAATRNLRRLLPSRRDYADLPRSWRRDVLAGVTVGVVALPLA
MSMEG_6453|M.smegmatis_MC2_155 --------MLAN--LARLLPHKNDYTDVPRTWRQDVLAGITVGVVALPLA
TH_2104|M.thermoresistible__bu ------VIATAKENLIRLLPSRRDYAELPRSWRRDILAGITVGVVALPLA
Mb1734|M.bovis_AF2122/97 ---------------------------MLQRIARELLSGVAVAIVALPLA
Rv1707|M.tuberculosis_H37Rv ---------------------------MLQRIARELLSGVAVAIVALPLA
MMAR_4750|M.marinum_M ---------------------MLAALRSPTRLRTEVLAGLVVALALIPEA
MUL_0321|M.ulcerans_Agy99 MSSQQVSAVPTIRKAGDPSTAVLAALRSPTRLRTEVLAGLVVALALIPEA
MLBr_02279|M.leprae_Br4923 ---------------------------MRSAIRYDLPASLVVFLVALPLS
MAV_0121|M.avium_104 -------------------MSTSRSERLREVLRYDLPASLVVFLVALPLS
MAB_4448|M.abscessus_ATCC_1997 ----------------MPSAPGLFESYDAIKARRDVIAGLTVAAISLPQA
:: :.:.* :* :
Mflv_1119|M.gilvum_PYR-GCK LAFGISSG---VGAAAGLITAVVAGLVAAVFGGSSVQVSGPTGAMAVVLA
Mvan_5693|M.vanbaalenii_PYR-1 LAFGISSG---VGAAAGLITAVVAGLVAAVFGGSAVQVSGPTGAMAVVLA
MSMEG_6453|M.smegmatis_MC2_155 LAFGISSG---VGAAAGLITAVVAGLVAAVFGGSHVQVSGPTGAMAVVLA
TH_2104|M.thermoresistible__bu LAFGISSG---VGAAAGLITAVVAGLVAAVFGGSHIQVSGPTGAMAVILA
Mb1734|M.bovis_AF2122/97 IAFGITATGTSQGALIGLYGAIFAGFFAAVFGGTPGQVTGPTGPITVVAT
Rv1707|M.tuberculosis_H37Rv IAFGITATGTSQGALIGLYGAIFAGFFAAVFGGTPGQVTGPTGPITVVAT
MMAR_4750|M.marinum_M ISFSIIAG---VDPRVGLFASFTMAVTIAVVGGRPAMISAATGAIALVIA
MUL_0321|M.ulcerans_Agy99 ISFSIIAG---VDPRVGLFASFTMAVTIAVVGGRLAMISAATGAIALVIA
MLBr_02279|M.leprae_Br4923 LGIAIASD---TPVLAGLIAAIVGGIVAGALGGSPLQVSGPAAGLTVVIA
MAV_0121|M.avium_104 LGIAIASD---APVLAGLIAAIVGGIVGGWLGGSPLQVSGPAAGLTVVVA
MAB_4448|M.abscessus_ATCC_1997 MAYALIAG---VDPKYGVYSAIVVTAIASIFGSSSHLINGPTSAISLLVF
:. .: : *: :. . .*. :...:. ::::
Mflv_1119|M.gilvum_PYR-GCK PIVAMHG-------LGSIALVTVLAGLLVLLAGVTGLGRAVTFIPWPVIE
Mvan_5693|M.vanbaalenii_PYR-1 PIVASHG-------LGSIALVTLLGGLIVLLAGVTGLGRAVTFIPWPVIE
MSMEG_6453|M.smegmatis_MC2_155 PIVAEHG-------LGSIALVTVLAGLLVLAAGITGLGRAVTFIPWPVIE
TH_2104|M.thermoresistible__bu PIVAQHG-------LTSIALVTVLAGLIVLAAGITGLGRAVTFIPWPVIE
Mb1734|M.bovis_AF2122/97 ATIAEHG-------LEGAFFAFILAGVFQILFGACRLGSLIRYVPHPVIS
Rv1707|M.tuberculosis_H37Rv ATIAEHG-------LEGAFFAFILAGVFQILFGACRLGSLIRYVPHPVIS
MMAR_4750|M.marinum_M PLAREHG-------ADYFIAAVILAGLVQITLSLLGVARLMRFIPRSVMV
MUL_0321|M.ulcerans_Agy99 PLAREQG-------ADYFIAAVILAGLVQITLSLLGVARLMRFIPRSVMV
MLBr_02279|M.leprae_Br4923 DLISDFG-------WGVTCFITVAAGVLQVLLGLSRVARAALAISPVVVH
MAV_0121|M.avium_104 DVVAEFG-------WGVTCFITVVAGVLQVLLGFSRIARAALAISPVVVH
MAB_4448|M.abscessus_ATCC_1997 SSLAFLDPENRTGLFEALFLLGVLVGTIQILIAVFKLGDLTRYISESVVI
. : * . : . :. :. *:
Mflv_1119|M.gilvum_PYR-GCK GFTLGIAAIIFLQQIPAAFAAGGPPGERTLPAAWHVVTHAD-----WSEA
Mvan_5693|M.vanbaalenii_PYR-1 GFTLGIAAIIFLQQVPAAFSATSPPGQRTLTAAWQVVTHAD-----WSQA
MSMEG_6453|M.smegmatis_MC2_155 GFTLGIAAIIFLQQVPAALAQQAPSGERTLLAAFTVVRNAD-----WGEA
TH_2104|M.thermoresistible__bu GFTLGIAAIIFLQQVPAAFATEAPAGQRTLVAAWHVVTHAD-----WTAA
Mb1734|M.bovis_AF2122/97 GFMGGIAILIIMTQLDQVRSS-------SLLVLVTVVLLLA-----SGRF
Rv1707|M.tuberculosis_H37Rv GFMGGIAILIIMTQLDQVRSS-------SLLVLVTVVLLLA-----SGRF
MMAR_4750|M.marinum_M GFVNALAILIFLSQLPHLLGVP----------------------------
MUL_0321|M.ulcerans_Agy99 GFVNALAILIFLSQVPHLLGVP----------------------------
MLBr_02279|M.leprae_Br4923 AMLAGVGITIALQQAHVLLGGKSKSTAWHNAIGLPSQILGA-----HRPG
MAV_0121|M.avium_104 AMLAGIGITIALQQVHVLLGGSSKSSAWSNVTGLPAQILGA-----HRPG
MAB_4448|M.abscessus_ATCC_1997 GFMASAALLLAIGQLANAIGVRDKGDGHMQVLQRAWLTLFHGDAVNYRAL
.: . . : : * .
Mflv_1119|M.gilvum_PYR-GCK VKSLGVIAFVVALMVALPRIHRAIPESLAAVVGATVAVGVLGISVATIG-
Mvan_5693|M.vanbaalenii_PYR-1 GKTLGVIAFVAALMLLLPRIHRAIPESLAAVIAATVVVAALGISVATIG-
MSMEG_6453|M.smegmatis_MC2_155 AKTLAVVGVVAVLMIVLPRVHSGIPASLTAVIVATVLIVALDIPVATIG-
TH_2104|M.thermoresistible__bu VKTLSVVSLVALLMVVLPRLHRSIPESLTAVVAATVIVTVFGISVATIG-
Mb1734|M.bovis_AF2122/97 IKAIPPSLLVLVLVSSVLPLAAPWLRDLRAGPVS------INRTVDYIG-
Rv1707|M.tuberculosis_H37Rv IKAIPPSLLVLVLVSSVLPLAAPWLRDLRAGPVS------INRTVDYIG-
MMAR_4750|M.marinum_M PVVYPFVIVGLLLMVFLPKLTTVVPAPLVAIVVLTAAAVAFSVEIPNVGD
MUL_0321|M.ulcerans_Agy99 AVVYPFVIVGLLLMVFLPKLTTVVPAPLVAIVVLTAAAVAFSVEILNVGD
MLBr_02279|M.leprae_Br4923 VILGVLVITILIAWRWVPARVARIPGPLVAIVVTTLVSVVFPFHVSRIQ-
MAV_0121|M.avium_104 LVLGLLVIAILVAWRWVPARLAIVPGPLVAIVVVTIISMVLPFKVSRIE-
MAB_4448|M.abscessus_ATCC_1997 FLSVTAVALAVLLRRLVQRYELPQIDMLLVLIVTAVIAYAYGWTVPDGTG
: * . :
Mflv_1119|M.gilvum_PYR-GCK --------ELPSHLPAPMLPQI--VPGALRDLFGAALAIAALAAIESLLS
Mvan_5693|M.vanbaalenii_PYR-1 --------ELPSHLPAPVLPHA--DAGALRDLFGAALAIAALAAIESLLS
MSMEG_6453|M.smegmatis_MC2_155 --------ELPSHLPAPVLPHV--DVNTLRTMFSAALAIAALAAIESLLS
TH_2104|M.thermoresistible__bu --------ELPSQLPTPVLPQA--DMGAMQTLLGAALAIAALAAIESLLS
Mb1734|M.bovis_AF2122/97 --------EIPQAMPSFDFPQV--ANSTMLQVLLSAVAIALLGSLDSLLT
Rv1707|M.tuberculosis_H37Rv --------EIPQAMPSFDFPQV--ANSTMLQVLLSAVAIALLGSLDSLLT
MMAR_4750|M.marinum_M E------GQLPSSLPTLLVPHVPFALETLSIIGPYALAMALVGLLESLLT
MUL_0321|M.ulcerans_Agy99 E------GQLPSSLPTLLVPHVPFALETLSIIGPYALAMALVGLLESLLT
MLBr_02279|M.leprae_Br4923 -------VDNRSSLDVLQLPKLP--DGHWGAVVLGVITVALIASVESLLS
MAV_0121|M.avium_104 -------LDG-SVLDAVRLPSLP--HGNWGAVAIAVITVTLITSVQSLLT
MAB_4448|M.abscessus_ATCC_1997 HTDVKISGKIPASLPEFHIPEVQ--VSTLGELSHGALAIAFIGLIEALSI
. : .* : .:::: : :::*
Mflv_1119|M.gilvum_PYR-GCK ARVAATMAPTGPYDPDRELVGQGLASVASGAFGGMPATGAIARTAVNVRS
Mvan_5693|M.vanbaalenii_PYR-1 ARVAATMAPTGPYDPDRELVGQGLASVASGVFGGMPATGAIARTAVNVRS
MSMEG_6453|M.smegmatis_MC2_155 ARVAATMSPTGPYDPNRELVGQGLASVASGAFGGMPATGAIARTAVNVRS
TH_2104|M.thermoresistible__bu ARVAATMSATGPYHPDRELVGQGMASVASGLFGGMPATGAIARTAVNVRS
Mb1734|M.bovis_AF2122/97 SLVMDNIRGTR-HRSNKELIGQGIGNIAAGLFGGLAGAGATVRSVVNVRN
Rv1707|M.tuberculosis_H37Rv SLVMDNIRGTR-HRSNKELIGQGIGNIAAGLFGGLSGAGATVRSVVNVRN
MMAR_4750|M.marinum_M AKLVDDITDTHSNKTR-EAWGQGVANVVTGFFGGMGGCAMIGQTMINVKV
MUL_0321|M.ulcerans_Agy99 AKLVDDITDTHSNKTR-EAWGQGVANVVTGFFGGMGGCAMIGQTMINVKV
MLBr_02279|M.leprae_Br4923 AVSLDKMHNGPRTDLNRELIGQGAANIVSGAIGGLPITGVIVRSSTNVIA
MAV_0121|M.avium_104 AVSIDRMHTGPRTDFNRELIGQGAANIASGALGGLPIAGVIVRSSANVNA
MAB_4448|M.abscessus_ATCC_1997 AKAIA-HHTGQQIDYNRQILAEGLANLAGGFFQSLPGSGSLSRSAINYQS
: : .:* ..:. * : .: . :: *
Mflv_1119|M.gilvum_PYR-GCK -GARTRLAAVVHSVVLLGVVYLATGPVAAIPLAALSGVLMVTSFRMISRS
Mvan_5693|M.vanbaalenii_PYR-1 -GARTRLSAVVHSLVLLGVVYLATGPVSAIPLAALSGVLMVTSFRMISWS
MSMEG_6453|M.smegmatis_MC2_155 -GAATRLSAIVHSLVLLAVVYLATGPVSAIPLSALAGVLMVTSFRMISAT
TH_2104|M.thermoresistible__bu -GARTRLAAIVHSLVLLAVVYLATGPVSAIPLSALAGVLMVTSFRMVSLH
Mb1734|M.bovis_AF2122/97 -GGQTALSAATHSVVLFVFVAGLGAVVQYIPLAVLSGILILVAVGMFDWH
Rv1707|M.tuberculosis_H37Rv -GGQTALSAATHSVVLFVFVAGLGAVVQYIPLAVLSGILILVAVGMFDWH
MMAR_4750|M.marinum_M SGARTRISTFLAGLFLLGLVVGLGDVVALIPMAALVAVMIMVSVGTMDWH
MUL_0321|M.ulcerans_Agy99 PGARTRISTFLAGLFLLGLVVGLGDVVALIPMAALVAVMIMVSVGTMDWH
MLBr_02279|M.leprae_Br4923 -GAKSRASSIMHGIWILLFTIPFAGLVDQIPSAALAGLLIVVGVQLLKPA
MAV_0121|M.avium_104 -GAKTRASTIMHGFWVLVFAVPFAGLVEKIPTAALAGLLIVIGIELLKPA
MAB_4448|M.abscessus_ATCC_1997 -GAATRFSGIVSAATVTIALLLFAPLLRYIPQAALAGLLLVTAVRLVDFH
*. : : . : : ** :.* .:::: .. ..
Mflv_1119|M.gilvum_PYR-GCK TMR--KILTSTRSDATTFVLTAVVTVCFD-LIEAVQIGILVAAFFALRSV
Mvan_5693|M.vanbaalenii_PYR-1 TIR--KIVTSTRSDALTFVLTAVITVCFD-LIEAVEIGILVAAFFALHSV
MSMEG_6453|M.smegmatis_MC2_155 TIR--KIVRSTRSDALIFVLTAVITVCFD-LIEAVEIGILVAAFFALRSV
TH_2104|M.thermoresistible__bu TAR--RILRSTRSDALTFVLTALVTITFD-LIEAVQIGVLVAAFFALRNV
Mb1734|M.bovis_AF2122/97 AMR--KAHVSPRGDVIVMFTTMIITVVVD-LTIAVMVGIALSLLVHRLRS
Rv1707|M.tuberculosis_H37Rv AMR--KAHVSPRGDVIVMFTTMIITVVVD-LTIAVMVGIALSLLVHRLRS
MMAR_4750|M.marinum_M SIAPATLRRMPRSETLVMLATVVVTVATHNLAYGVVAGVLTAMVLFARRV
MUL_0321|M.ulcerans_Agy99 SIAPATLRRMPRSETLVMLATVVVTVATHNLAYGVVAGVLTAMVLFARRV
MLBr_02279|M.leprae_Br4923 HIET-----ALRNGDLAVYLVTAVCVVFLNLLHGVMIGLAVAIALTGWRV
MAV_0121|M.avium_104 HIET-----ALRNGDLAIYLVTVTSVIFLNLLHGVLIGLLLAVVVTGWRV
MAB_4448|M.abscessus_ATCC_1997 RLVY--AIKASRYDAGLVIITALVGVAVN-LDTAVLVGVVLSILLFVPRA
* . . : * .* *: : .
Mflv_1119|M.gilvum_PYR-GCK ARRSS---------VVREELPAPHRPGDEEIALLRLDGAMFFGAAERISN
Mvan_5693|M.vanbaalenii_PYR-1 ARRSS---------VVREELPAPYQPGDEEIALLRLDGAMFFGVAERISN
MSMEG_6453|M.smegmatis_MC2_155 ARRSS---------VVREELPGPRRPGDDQIALLRLDGAMFFGAAERISN
TH_2104|M.thermoresistible__bu ARRSG---------VTREELPGEPQPGDERIALLRLEGSMFFGVAERMSS
Mb1734|M.bovis_AF2122/97 RQRKA---------KVTQDDTG----------TYRIDGPLSFLSVDGVFG
Rv1707|M.tuberculosis_H37Rv RQRKA---------KVTQDDTG----------TYRIDGPLSFLSVDGVFG
MMAR_4750|M.marinum_M AHFMT---------VERIVESDTG--SAAETVVYRVSGELFFASSNDLVY
MUL_0321|M.ulcerans_Agy99 AHFMT---------VERIVESDTG--SAAETVVYRVSCELFFASSNDLVY
MLBr_02279|M.leprae_Br4923 IRARV---------EAQPVGDEWH---------VLVAGACTFLALPRLTR
MAV_0121|M.avium_104 VRARI---------EAEPVGDGWH---------VVIEGACTFLALPRLTG
MAB_4448|M.abscessus_ATCC_1997 AKLKAAELVVTDEGVIRERTPQDRPTGAGAPVIYDLEGELFFGAAPELDR
: : * :
Mflv_1119|M.gilvum_PYR-GCK AIV----DGDHPRTSVVIIRMSQLGMLDATGANTLAQICTELEARGITVI
Mvan_5693|M.vanbaalenii_PYR-1 AII----DSDHPRTSVVIIRMSQLGMLDATGANTLAQICTELEARGITVI
MSMEG_6453|M.smegmatis_MC2_155 AIT----DAEHPHVSVVIIRMSQLGMLDATGAHTLAEITTELESRGITVI
TH_2104|M.thermoresistible__bu TIS----EIEADDISVVIIRLSQLGTLDATGANSLAQIATELERRGITVI
Mb1734|M.bovis_AF2122/97 SLR----DGRED----VSLDLQHVTYLDTSGAQALLYFIDHSEKDGVAVS
Rv1707|M.tuberculosis_H37Rv SLR----DGRED----VSLDLQHVTYLDTSGARALLYFIDHSEKDGVAVS
MMAR_4750|M.marinum_M QFN----YVDDPDN--VVIDMSQSHIWDASTVAALDAIITKYHAKGRCVE
MUL_0321|M.ulcerans_Agy99 QFN----YVDDPDN--VVIDMSQSHIWDASTVAALDAIITKYHAKGRCVE
MLBr_02279|M.leprae_Br4923 VLA----SVPPCTR---VTVNISVNYLDHAAHQAIHDWQRQHHATGGTVH
MAV_0121|M.avium_104 VLA----SIPERTS---VTVHLLTNYLDHAAHQAIGDWQRRHCATGGTVE
MAB_4448|M.abscessus_ATCC_1997 YLESLHTRIRDEHLKVVILRLKRVRHPDVVCIERLEHFIREQQQLGVTVL
: * : . * *
Mflv_1119|M.gilvum_PYR-GCK IKGVRPEHSALLANVGVFESLRHERHLVDTLDEAIEHARTHARRQPH---
Mvan_5693|M.vanbaalenii_PYR-1 IKGVRPEHSALLSSVGVFESLRHENHLVDTLDEAIEHARTHAHRQPH---
MSMEG_6453|M.smegmatis_MC2_155 IKGVRPEHMDLLANVGVFESLRHENHLLDSLDDAVAHARTHVARQPH---
TH_2104|M.thermoresistible__bu IKGVQPEHSKLLSNVGVFESLRHEKHLIDSLEDAIEHARSHVARQQTART
Mb1734|M.bovis_AF2122/97 IKRIPPRLESQLTALADNEQRDKLRTVLESA-------------------
Rv1707|M.tuberculosis_H37Rv IKRIPPRLESQLTALADNEQRDKLRTVLESA-------------------
MMAR_4750|M.marinum_M IIGLNQDSAERHGRLSGQFGAEH---------------------------
MUL_0321|M.ulcerans_Agy99 IIGLNQDSAERHGRLSGQFGAEH---------------------------
MLBr_02279|M.leprae_Br4923 INGVVETGHFSRCAMGDLIGPEQQPEAAA---------------------
MAV_0121|M.avium_104 VRDTTEPAARRRNSHLSLVEQVSSPGGA----------------------
MAB_4448|M.abscessus_ATCC_1997 LAGVRPDSLALLENVGFTHWLPPEQVFPEEEKEFSATLRAVRYAQSRLEE
:
Mflv_1119|M.gilvum_PYR-GCK ----------------
Mvan_5693|M.vanbaalenii_PYR-1 ----------------
MSMEG_6453|M.smegmatis_MC2_155 ----------------
TH_2104|M.thermoresistible__bu TS--------------
Mb1734|M.bovis_AF2122/97 ----------------
Rv1707|M.tuberculosis_H37Rv ----------------
MMAR_4750|M.marinum_M ----------------
MUL_0321|M.ulcerans_Agy99 ----------------
MLBr_02279|M.leprae_Br4923 ----------------
MAV_0121|M.avium_104 ----------------
MAB_4448|M.abscessus_ATCC_1997 EPPNPAANPEKLYYLV