For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ITIGRLGRLLPQRSDYTDLRRSWRADLLAGVTVGVVALPLALAFGISSGVGAAAGLITAVVAGLVAAVFG GSSVQVSGPTGAMAVVLAPIVAMHGLGSIALVTVLAGLLVLLAGVTGLGRAVTFIPWPVIEGFTLGIAAI IFLQQIPAAFAAGGPPGERTLPAAWHVVTHADWSEAVKSLGVIAFVVALMVALPRIHRAIPESLAAVVGA TVAVGVLGISVATIGELPSHLPAPMLPQIVPGALRDLFGAALAIAALAAIESLLSARVAATMAPTGPYDP DRELVGQGLASVASGAFGGMPATGAIARTAVNVRSGARTRLAAVVHSVVLLGVVYLATGPVAAIPLAALS GVLMVTSFRMISRSTMRKILTSTRSDATTFVLTAVVTVCFDLIEAVQIGILVAAFFALRSVARRSSVVRE ELPAPHRPGDEEIALLRLDGAMFFGAAERISNAIVDGDHPRTSVVIIRMSQLGMLDATGANTLAQICTEL EARGITVIIKGVRPEHSALLANVGVFESLRHERHLVDTLDEAIEHARTHARRQPH*
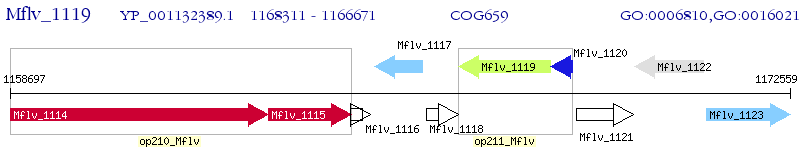
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1119 | - | - | 100% (546) | sulphate transporter |
| M. gilvum PYR-GCK | Mflv_4975 | - | 6e-47 | 28.65% (541) | sulfate transporter |
| M. gilvum PYR-GCK | Mflv_0346 | - | 4e-11 | 25.18% (421) | xanthine/uracil/vitamin C permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1734 | - | 3e-63 | 33.33% (498) | transmembrane protein |
| M. tuberculosis H37Rv | Rv1707 | - | 3e-63 | 33.33% (498) | transmembrane protein |
| M. leprae Br4923 | MLBr_02279 | - | 2e-51 | 32.66% (493) | putative transmembrane transport protein |
| M. abscessus ATCC 19977 | MAB_4448 | - | 7e-56 | 29.43% (564) | putative sulfate transporter |
| M. marinum M | MMAR_1266 | sulP_2 | 7e-55 | 32.46% (499) | transmembrane carbonic anhydrase, SulP_2 |
| M. avium 104 | MAV_4578 | - | 3e-53 | 32.42% (509) | carbonate dehydratase |
| M. smegmatis MC2 155 | MSMEG_6453 | - | 0.0 | 80.85% (543) | sulfate permease |
| M. thermoresistible (build 8) | TH_2104 | - | 0.0 | 79.48% (536) | sulfate permease |
| M. ulcerans Agy99 | MUL_0321 | - | 6e-51 | 32.14% (501) | integral membrane transporter |
| M. vanbaalenii PYR-1 | Mvan_5693 | - | 0.0 | 89.81% (540) | sulphate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1119|M.gilvum_PYR-GCK ------MITIGR--LGRLLPQRSDYTDLRRSWRADLLAGVTVGVVALPLA
Mvan_5693|M.vanbaalenii_PYR-1 ------MIAAATRNLRRLLPSRRDYADLPRSWRRDVLAGVTVGVVALPLA
MSMEG_6453|M.smegmatis_MC2_155 --------MLAN--LARLLPHKNDYTDVPRTWRQDVLAGITVGVVALPLA
TH_2104|M.thermoresistible__bu ------VIATAKENLIRLLPSRRDYAELPRSWRRDILAGITVGVVALPLA
Mb1734|M.bovis_AF2122/97 ---------------------------MLQRIARELLSGVAVAIVALPLA
Rv1707|M.tuberculosis_H37Rv ---------------------------MLQRIARELLSGVAVAIVALPLA
MLBr_02279|M.leprae_Br4923 ---------------------------MRSAIRYDLPASLVVFLVALPLS
MAV_4578|M.avium_104 --------MQDIQRADDALPTASRAERLRGVIRHDLPSSLVVFLVALPLS
MMAR_1266|M.marinum_M ---------MSTAVKNSAEAPESRSQWMRANLRHDFPASLVVFLVALPLS
MUL_0321|M.ulcerans_Agy99 MSSQQVSAVPTIRKAGDPSTAVLAALRSPTRLRTEVLAGLVVALALIPEA
MAB_4448|M.abscessus_ATCC_1997 ----------------MPSAPGLFESYDAIKARRDVIAGLTVAAISLPQA
:. :.:.* :* :
Mflv_1119|M.gilvum_PYR-GCK LAFGISSG---VGAAAGLITAVVAGLVAAVFGGSSVQVSGPTGAMAVVLA
Mvan_5693|M.vanbaalenii_PYR-1 LAFGISSG---VGAAAGLITAVVAGLVAAVFGGSAVQVSGPTGAMAVVLA
MSMEG_6453|M.smegmatis_MC2_155 LAFGISSG---VGAAAGLITAVVAGLVAAVFGGSHVQVSGPTGAMAVVLA
TH_2104|M.thermoresistible__bu LAFGISSG---VGAAAGLITAVVAGLVAAVFGGSHIQVSGPTGAMAVILA
Mb1734|M.bovis_AF2122/97 IAFGITATGTSQGALIGLYGAIFAGFFAAVFGGTPGQVTGPTGPITVVAT
Rv1707|M.tuberculosis_H37Rv IAFGITATGTSQGALIGLYGAIFAGFFAAVFGGTPGQVTGPTGPITVVAT
MLBr_02279|M.leprae_Br4923 LGIAIASD---TPVLAGLIAAIVGGIVAGALGGSPLQVSGPAAGLTVVIA
MAV_4578|M.avium_104 LGIAIASN---APVLAGLIAAIVGGIVVGALGGSPLQVSGPAAGLTVVVA
MMAR_1266|M.marinum_M LGIAIASG---APLMAGLIAAVVGGIVAGLLGGSSVQVSGPAAGLTVVVA
MUL_0321|M.ulcerans_Agy99 ISFSIIAG---VDPRVGLFASFTMAVTIAVVGGRLAMISAATGAIALVIA
MAB_4448|M.abscessus_ATCC_1997 MAYALIAG---VDPKYGVYSAIVVTAIASIFGSSSHLINGPTSAISLLVF
:. .: : *: :. . .*. :...:. ::::
Mflv_1119|M.gilvum_PYR-GCK PIVAMHG-------LGSIALVTVLAGLLVLLAGVTGLGRAVTFIPWPVIE
Mvan_5693|M.vanbaalenii_PYR-1 PIVASHG-------LGSIALVTLLGGLIVLLAGVTGLGRAVTFIPWPVIE
MSMEG_6453|M.smegmatis_MC2_155 PIVAEHG-------LGSIALVTVLAGLLVLAAGITGLGRAVTFIPWPVIE
TH_2104|M.thermoresistible__bu PIVAQHG-------LTSIALVTVLAGLIVLAAGITGLGRAVTFIPWPVIE
Mb1734|M.bovis_AF2122/97 ATIAEHG-------LEGAFFAFILAGVFQILFGACRLGSLIRYVPHPVIS
Rv1707|M.tuberculosis_H37Rv ATIAEHG-------LEGAFFAFILAGVFQILFGACRLGSLIRYVPHPVIS
MLBr_02279|M.leprae_Br4923 DLISDFG-------WGVTCFITVAAGVLQVLLGLSRVARAALAISPVVVH
MAV_4578|M.avium_104 GLVSDFG-------WGVTCFITVAAGAVQVLLGLSRVARAALAISPVVVH
MMAR_1266|M.marinum_M GLIEQVG-------WQMLCLMTLGAGVLQIIFGLSRMARAALAIAPVVVH
MUL_0321|M.ulcerans_Agy99 PLAREQG-------ADYFIAAVILAGLVQITLSLLGVARLMRFIPRSVMV
MAB_4448|M.abscessus_ATCC_1997 SSLAFLDPENRTGLFEALFLLGVLVGTIQILIAVFKLGDLTRYISESVVI
. : * . : . :. :. *:
Mflv_1119|M.gilvum_PYR-GCK GFTLGIAAIIFLQQIPAAFAAGGPPGERTLPAAWHVVTHAD-----WSEA
Mvan_5693|M.vanbaalenii_PYR-1 GFTLGIAAIIFLQQVPAAFSATSPPGQRTLTAAWQVVTHAD-----WSQA
MSMEG_6453|M.smegmatis_MC2_155 GFTLGIAAIIFLQQVPAALAQQAPSGERTLLAAFTVVRNAD-----WGEA
TH_2104|M.thermoresistible__bu GFTLGIAAIIFLQQVPAAFATEAPAGQRTLVAAWHVVTHAD-----WTAA
Mb1734|M.bovis_AF2122/97 GFMGGIAILIIMTQLDQVRSS-------SLLVLVTVVLLLA-----SGRF
Rv1707|M.tuberculosis_H37Rv GFMGGIAILIIMTQLDQVRSS-------SLLVLVTVVLLLA-----SGRF
MLBr_02279|M.leprae_Br4923 AMLAGVGITIALQQAHVLLGGKSKSTAWHNAIGLPSQILGA-----HRPG
MAV_4578|M.avium_104 AMLAGIGITIALQQTHVLLGGKSKSTAWHNLIGLPGQIIGA-----HRPG
MMAR_1266|M.marinum_M AMLAGIGVTIVLQQIHVLMGGSAHSSAWRNIAALPNGILHH-----ELHE
MUL_0321|M.ulcerans_Agy99 GFVNALAILIFLSQVPHLLGVP----------------------------
MAB_4448|M.abscessus_ATCC_1997 GFMASAALLLAIGQLANAIGVRDKGDGHMQVLQRAWLTLFHGDAVNYRAL
.: . . : : * .
Mflv_1119|M.gilvum_PYR-GCK VKSLGVIAFVVALMVALPRIHRAIPESLAAVVGATVAVGVLGISVAT---
Mvan_5693|M.vanbaalenii_PYR-1 GKTLGVIAFVAALMLLLPRIHRAIPESLAAVIAATVVVAALGISVAT---
MSMEG_6453|M.smegmatis_MC2_155 AKTLAVVGVVAVLMIVLPRVHSGIPASLTAVIVATVLIVALDIPVAT---
TH_2104|M.thermoresistible__bu VKTLSVVSLVALLMVVLPRLHRSIPESLTAVVAATVIVTVFGISVAT---
Mb1734|M.bovis_AF2122/97 IKAIPPSLLVLVLVSSVLPLAAPWLRDLRAGPVS------INRTVDY---
Rv1707|M.tuberculosis_H37Rv IKAIPPSLLVLVLVSSVLPLAAPWLRDLRAGPVS------INRTVDY---
MLBr_02279|M.leprae_Br4923 VILGVLVITILIAWRWVPARVARIPGPLVAIVVTTLVSVVFPFHVSR---
MAV_4578|M.avium_104 VLLGVLVIVILVAWRWVPAKVRRVPGPLVAIVAVTVISVVFPFHVRR---
MMAR_1266|M.marinum_M VIVGGTVIAILLVWSRLPAMVRMVPGPLVAIAGATALSLMVNLDVER---
MUL_0321|M.ulcerans_Agy99 AVVYPFVIVGLLLMVFLPKLTTVVPAPLVAIVVLTAAAVAFSVEILN---
MAB_4448|M.abscessus_ATCC_1997 FLSVTAVALAVLLRRLVQRYELPQIDMLLVLIVTAVIAYAYGWTVPDGTG
: * . :
Mflv_1119|M.gilvum_PYR-GCK ---IGELP---SHLPAPMLPQIVPGA---------LRDLFGAALAIAALA
Mvan_5693|M.vanbaalenii_PYR-1 ---IGELP---SHLPAPVLPHADAGA---------LRDLFGAALAIAALA
MSMEG_6453|M.smegmatis_MC2_155 ---IGELP---SHLPAPVLPHVDVNT---------LRTMFSAALAIAALA
TH_2104|M.thermoresistible__bu ---IGELP---SQLPTPVLPQADMGA---------MQTLLGAALAIAALA
Mb1734|M.bovis_AF2122/97 ---IGEIP---QAMPSFDFPQVANST---------MLQVLLSAVAIALLG
Rv1707|M.tuberculosis_H37Rv ---IGEIP---QAMPSFDFPQVANST---------MLQVLLSAVAIALLG
MLBr_02279|M.leprae_Br4923 ---IQVDNR--SSLDVLQLPKLPDGH-----WG----AVVLGVITVALIA
MAV_4578|M.avium_104 ---IDLDG---SPLDALQLPDLPHGN-----WS----GVAVGVITVALIA
MMAR_1266|M.marinum_M ---INLSG---NFFEAISLPRLPELTPDGQPWSNDISIIAVGVLTIALIA
MUL_0321|M.ulcerans_Agy99 ---VGDEGQLPSSLPTLLVPHVPFALET-------LSIIGPYALAMALVG
MAB_4448|M.abscessus_ATCC_1997 HTDVKISGKIPASLPEFHIPEVQVST---------LGELSHGALAIAFIG
: : .* : .:::* :.
Mflv_1119|M.gilvum_PYR-GCK AIESLLSARVAATMAPTGPYDPDRELVGQGLASVASGAFGGMPATGAIAR
Mvan_5693|M.vanbaalenii_PYR-1 AIESLLSARVAATMAPTGPYDPDRELVGQGLASVASGVFGGMPATGAIAR
MSMEG_6453|M.smegmatis_MC2_155 AIESLLSARVAATMSPTGPYDPNRELVGQGLASVASGAFGGMPATGAIAR
TH_2104|M.thermoresistible__bu AIESLLSARVAATMSATGPYHPDRELVGQGMASVASGLFGGMPATGAIAR
Mb1734|M.bovis_AF2122/97 SLDSLLTSLVMDNIRGTR-HRSNKELIGQGIGNIAAGLFGGLAGAGATVR
Rv1707|M.tuberculosis_H37Rv SLDSLLTSLVMDNIRGTR-HRSNKELIGQGIGNIAAGLFGGLSGAGATVR
MLBr_02279|M.leprae_Br4923 SVESLLSAVSLDKMHNGPRTDLNRELIGQGAANIVSGAIGGLPITGVIVR
MAV_4578|M.avium_104 SVESLLSAVSVDRMHNGPRTDFNRELVGQGAANMISGAVGGLPVTGVIVR
MMAR_1266|M.marinum_M SVESLLCAVGVDKLHHGPRTNFNREMLGQGSANVLSGLLGGLPVTGVIVR
MUL_0321|M.ulcerans_Agy99 LLESLLTAKLVDDITDTHSNKTR-EAWGQGVANVVTGFFGGMGGCAMIGQ
MAB_4448|M.abscessus_ATCC_1997 LIEALSIAKAIA-HHTGQQIDYNRQILAEGLANLAGGFFQSLPGSGSLSR
:::* : : .:* ..: * . .: . :
Mflv_1119|M.gilvum_PYR-GCK TAVNVRS-GARTRLAAVVHSVVLLGVVYLATGPVAAIPLAALSGVLMVTS
Mvan_5693|M.vanbaalenii_PYR-1 TAVNVRS-GARTRLSAVVHSLVLLGVVYLATGPVSAIPLAALSGVLMVTS
MSMEG_6453|M.smegmatis_MC2_155 TAVNVRS-GAATRLSAIVHSLVLLAVVYLATGPVSAIPLSALAGVLMVTS
TH_2104|M.thermoresistible__bu TAVNVRS-GARTRLAAIVHSLVLLAVVYLATGPVSAIPLSALAGVLMVTS
Mb1734|M.bovis_AF2122/97 SVVNVRN-GGQTALSAATHSVVLFVFVAGLGAVVQYIPLAVLSGILILVA
Rv1707|M.tuberculosis_H37Rv SVVNVRN-GGQTALSAATHSVVLFVFVAGLGAVVQYIPLAVLSGILILVA
MLBr_02279|M.leprae_Br4923 SSTNVIA-GAKSRASSIMHGIWILLFTIPFAGLVDQIPSAALAGLLIVVG
MAV_4578|M.avium_104 SSTNVNA-GARSRASAIMHGVWILLFTIPFAGLVDEIPTAALAGLLIVIG
MMAR_1266|M.marinum_M SSANVAA-GARTRTSAVLHGVWILLFASLFTGLVELIPKAALAGLLIVVG
MUL_0321|M.ulcerans_Agy99 TMINVKVPGARTRISTFLAGLFLLGLVVGLGDVVALIPMAALVAVMIMVS
MAB_4448|M.abscessus_ATCC_1997 SAINYQS-GAATRFSGIVSAATVTIALLLFAPLLRYIPQAALAGLLLVTA
: * *. : : . : : ** :.* .:::: .
Mflv_1119|M.gilvum_PYR-GCK FRMISRSTMR--KILTSTRSDATTFVLTAVVTVCFD-LIEAVQIGILVAA
Mvan_5693|M.vanbaalenii_PYR-1 FRMISWSTIR--KIVTSTRSDALTFVLTAVITVCFD-LIEAVEIGILVAA
MSMEG_6453|M.smegmatis_MC2_155 FRMISATTIR--KIVRSTRSDALIFVLTAVITVCFD-LIEAVEIGILVAA
TH_2104|M.thermoresistible__bu FRMVSLHTAR--RILRSTRSDALTFVLTALVTITFD-LIEAVQIGVLVAA
Mb1734|M.bovis_AF2122/97 VGMFDWHAMR--KAHVSPRGDVIVMFTTMIITVVVD-LTIAVMVGIALSL
Rv1707|M.tuberculosis_H37Rv VGMFDWHAMR--KAHVSPRGDVIVMFTTMIITVVVD-LTIAVMVGIALSL
MLBr_02279|M.leprae_Br4923 VQLLKPAHIE--TALRN--GDLAVYLVTAVCVVFLN-LLHGVMIGLAVAI
MAV_4578|M.avium_104 IQLLKPAHIE--TAMKH--GDLAVYVVTAVSVIFLN-LLHGVMIGLALAI
MMAR_1266|M.marinum_M AQLIQLAHIQ--IAWRT--GNFAIYVITILGVIFLN-LLEGVVIGLGVAI
MUL_0321|M.ulcerans_Agy99 VGTMDWHSIAPATLRRMPRSETLVMLATVVVTVATHNLAYGVVAGVLTAM
MAB_4448|M.abscessus_ATCC_1997 VRLVDFHRLVY--AIKASRYDAGLVIITALVGVAVN-LDTAVLVGVVLSI
.. : . * : : . * .* *: :
Mflv_1119|M.gilvum_PYR-GCK FFALRSVARRSS---------VVREELPAPHRPGDEEIALLRLDG-AMFF
Mvan_5693|M.vanbaalenii_PYR-1 FFALHSVARRSS---------VVREELPAPYQPGDEEIALLRLDG-AMFF
MSMEG_6453|M.smegmatis_MC2_155 FFALRSVARRSS---------VVREELPGPRRPGDDQIALLRLDG-AMFF
TH_2104|M.thermoresistible__bu FFALRNVARRSG---------VTREELPGEPQPGDERIALLRLEG-SMFF
Mb1734|M.bovis_AF2122/97 LVHRLRSRQRKA---------KVTQDDTG----------TYRIDG-PLSF
Rv1707|M.tuberculosis_H37Rv LVHRLRSRQRKA---------KVTQDDTG----------TYRIDG-PLSF
MLBr_02279|M.leprae_Br4923 ALTGWRVIRARV---------EAQPVGDE---------WHVLVAG-ACTF
MAV_4578|M.avium_104 ALTGWRVIRAKI---------EAAQLDGE---------WRVTIEGAACTF
MMAR_1266|M.marinum_M AFLLIRVIRAPI---------EARLVSEEAKR------WRVDIDG-TLSF
MUL_0321|M.ulcerans_Agy99 VLFARRVAHFMT---------VERIVESDTGS---AAETVVYRVSCELFF
MAB_4448|M.abscessus_ATCC_1997 LLFVPRAAKLKAAELVVTDEGVIRERTPQDRPTGAGAPVIYDLEG-ELFF
. : . *
Mflv_1119|M.gilvum_PYR-GCK GAAERISNAIV----DGDHPRTSVVIIRMSQLGMLDATGANTLAQICTEL
Mvan_5693|M.vanbaalenii_PYR-1 GVAERISNAII----DSDHPRTSVVIIRMSQLGMLDATGANTLAQICTEL
MSMEG_6453|M.smegmatis_MC2_155 GAAERISNAIT----DAEHPHVSVVIIRMSQLGMLDATGAHTLAEITTEL
TH_2104|M.thermoresistible__bu GVAERMSSTIS----EIEADDISVVIIRLSQLGTLDATGANSLAQIATEL
Mb1734|M.bovis_AF2122/97 LSVDGVFGSLR----DGRED----VSLDLQHVTYLDTSGAQALLYFIDHS
Rv1707|M.tuberculosis_H37Rv LSVDGVFGSLR----DGRED----VSLDLQHVTYLDTSGARALLYFIDHS
MLBr_02279|M.leprae_Br4923 LALPRLTRVLA----SVPPC---TRVTVNISVNYLDHAAHQAIHDWQRQH
MAV_4578|M.avium_104 LALPRLTRVLA----SVPRG---ATVTVAIAVHYLDHAAHQAITDWQRQQ
MMAR_1266|M.marinum_M LLLPRLTNVLS----ELPEG---SDITLNLNADYIDHSVSEAISDWKTAH
MUL_0321|M.ulcerans_Agy99 ASSNDLVYQFN----YVDDPD--NVVIDMSQSHIWDASTVAALDAIITKY
MAB_4448|M.abscessus_ATCC_1997 GAAPELDRYLESLHTRIRDEHLKVVILRLKRVRHPDVVCIERLEHFIREQ
: : * :
Mflv_1119|M.gilvum_PYR-GCK EARGITVIIKGVRP------------------------------------
Mvan_5693|M.vanbaalenii_PYR-1 EARGITVIIKGVRP------------------------------------
MSMEG_6453|M.smegmatis_MC2_155 ESRGITVIIKGVRP------------------------------------
TH_2104|M.thermoresistible__bu ERRGITVIIKGVQP------------------------------------
Mb1734|M.bovis_AF2122/97 EKDGVAVSIKRIPP------------------------------------
Rv1707|M.tuberculosis_H37Rv EKDGVAVSIKRIPP------------------------------------
MLBr_02279|M.leprae_Br4923 HATGGTVHING---------------------------------------
MAV_4578|M.avium_104 EATGGTVRIEGA--------------------------------------
MMAR_1266|M.marinum_M EATGGEVTIIETSPANLVSAHTSPPKRHFMSRALREPWTSRRDSHPDAEP
MUL_0321|M.ulcerans_Agy99 HAKGRCVEIIGLNQ------------------------------------
MAB_4448|M.abscessus_ATCC_1997 QQLGVTVLLAGVRP------------------------------------
. * * :
Mflv_1119|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5693|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6453|M.smegmatis_MC2_155 --------------------------------------------------
TH_2104|M.thermoresistible__bu --------------------------------------------------
Mb1734|M.bovis_AF2122/97 --------------------------------------------------
Rv1707|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02279|M.leprae_Br4923 --------------------------------------------------
MAV_4578|M.avium_104 --------------------------------------------------
MMAR_1266|M.marinum_M SIRHGVEEYHRNGIGTLHHHVRELMDPANPDTVFLTCADSRILPDVITAS
MUL_0321|M.ulcerans_Agy99 --------------------------------------------------
MAB_4448|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1119|M.gilvum_PYR-GCK ----------------------------------EHSALLANVGVFESLR
Mvan_5693|M.vanbaalenii_PYR-1 ----------------------------------EHSALLSSVGVFESLR
MSMEG_6453|M.smegmatis_MC2_155 ----------------------------------EHMDLLANVGVFESLR
TH_2104|M.thermoresistible__bu ----------------------------------EHSKLLSNVGVFESLR
Mb1734|M.bovis_AF2122/97 ----------------------------------RLESQLTALADNEQRD
Rv1707|M.tuberculosis_H37Rv ----------------------------------RLESQLTALADNEQRD
MLBr_02279|M.leprae_Br4923 ---------------------------------------VVETGHFSRCA
MAV_4578|M.avium_104 ---------------------------------------VVATGRRD---
MMAR_1266|M.marinum_M KPGDLYIVRNVGNLVPIDPTERSVDAALDFAVNQLGVSSVVVCGHSSCRS
MUL_0321|M.ulcerans_Agy99 -------------------------------------DSAERHGRLSGQF
MAB_4448|M.abscessus_ATCC_1997 ----------------------------------DSLALLENVGFTHWLP
.
Mflv_1119|M.gilvum_PYR-GCK HERHLVDTLDEAIEHAR-----------THARRQPH--------------
Mvan_5693|M.vanbaalenii_PYR-1 HENHLVDTLDEAIEHAR-----------THAHRQPH--------------
MSMEG_6453|M.smegmatis_MC2_155 HENHLLDSLDDAVAHAR-----------THVARQPH--------------
TH_2104|M.thermoresistible__bu HEKHLIDSLEDAIEHAR-----------SHVARQQTARTTS---------
Mb1734|M.bovis_AF2122/97 KLRTVLESA-----------------------------------------
Rv1707|M.tuberculosis_H37Rv KLRTVLESA-----------------------------------------
MLBr_02279|M.leprae_Br4923 MGDLIGP------------------------EQQPEAAA-----------
MAV_4578|M.avium_104 --EPQVE------------------------AEMPGAAA-----------
MMAR_1266|M.marinum_M MQALLDNGASDVDRPMNHWLEHAHDSLAAFRDGHPARASAASVGFGELDQ
MUL_0321|M.ulcerans_Agy99 GAEH----------------------------------------------
MAB_4448|M.abscessus_ATCC_1997 PEQVFPEEEKEFSATLRAVRYAQSRLEEEPPNPAANPEKLYYLV------
Mflv_1119|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5693|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6453|M.smegmatis_MC2_155 --------------------------------------------------
TH_2104|M.thermoresistible__bu --------------------------------------------------
Mb1734|M.bovis_AF2122/97 --------------------------------------------------
Rv1707|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02279|M.leprae_Br4923 --------------------------------------------------
MAV_4578|M.avium_104 --------------------------------------------------
MMAR_1266|M.marinum_M LAVVNVAVQLERLAHNQVLAPAIASGAIQIVGMFFDFSTVHVHEVDQLGI
MUL_0321|M.ulcerans_Agy99 --------------------------------------------------
MAB_4448|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1119|M.gilvum_PYR-GCK ----------
Mvan_5693|M.vanbaalenii_PYR-1 ----------
MSMEG_6453|M.smegmatis_MC2_155 ----------
TH_2104|M.thermoresistible__bu ----------
Mb1734|M.bovis_AF2122/97 ----------
Rv1707|M.tuberculosis_H37Rv ----------
MLBr_02279|M.leprae_Br4923 ----------
MAV_4578|M.avium_104 ----------
MMAR_1266|M.marinum_M VASNQPVGKP
MUL_0321|M.ulcerans_Agy99 ----------
MAB_4448|M.abscessus_ATCC_1997 ----------