For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGGRPTRALPLLRTLLFVPGDRPDRIAKAAASAAAGVAVDLEDAVAVSRKDAARDAAVTALSALPTAGVP VLCVRINAVDSGFAEDDLTALEPVLHRLDLVMVPMSAEPDRIRRVSELLTRAERSAGADDGRIGIIPLVE TAAGILNAPSLASADARVHTLSFGPADLSRELGITPTSDGDEFLFARSQLVLAAAAANCPAPVDGPYLDL DDDDGLIRSARHARKLGFGGKQVIHPSQIASVAGAFVTSDAEIDWARAVDAAFSAAEAEGVSSIRLADGT FVDYPVAQRARAILAGAR
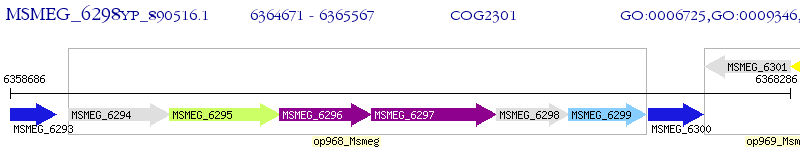
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6298 | - | - | 100% (298) | malyl-CoA lyase |
| M. smegmatis MC2 155 | MSMEG_5064 | - | 4e-27 | 31.65% (297) | malyl-CoA lyase |
| M. smegmatis MC2 155 | MSMEG_6644 | - | 2e-25 | 30.54% (298) | malyl-CoA lyase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2526c | citE | 1e-23 | 37.15% (288) | citrate (Pro-3S)-lyase beta subunit |
| M. gilvum PYR-GCK | Mflv_2556 | - | 3e-27 | 35.31% (286) | HpcH/HpaI aldolase |
| M. tuberculosis H37Rv | Rv2498c | citE | 1e-23 | 37.15% (288) | citrate (Pro-3S)-lyase beta subunit |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1369 | - | 6e-29 | 35.69% (255) | citrate lyase beta chain |
| M. marinum M | MMAR_4158 | citE_1 | 5e-30 | 34.26% (289) | citrate (pro-3s)-lyase (beta subunit) CitE_1 |
| M. avium 104 | MAV_5074 | - | 3e-41 | 40.56% (286) | citrate lyase |
| M. thermoresistible (build 8) | TH_0719 | - | 1e-23 | 34.39% (285) | HpcH/HpaI aldolase/citrate lyase family protein |
| M. ulcerans Agy99 | MUL_4021 | citE_1 | 2e-30 | 34.60% (289) | citrate (pro-3s)-lyase (beta subunit) CitE_1 |
| M. vanbaalenii PYR-1 | Mvan_4487 | - | 4e-26 | 32.21% (298) | citrate (pro-3S)-lyase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6298|M.smegmatis_MC2_155 MGGRPTRALPLLRTLLFVPGDRPDRIAKAAASAAAGVAVDLEDAVAVSRK
MAV_5074|M.avium_104 ----MHDDLASARSLLFVPGDRPDRFTKAASSDADGIIIDLEDAVAPEAK
MMAR_4158|M.marinum_M -------MKPIPRSLLFTPATKTQHYLTAAAVHAEALILDLEDSVAPRDK
MUL_4021|M.ulcerans_Agy99 -------MKPIPRSLLFTPATKTQHYLTAAAVHAEALILDLEDSVAPRDK
Mb2526c|M.bovis_AF2122/97 -----MNLRAAGPGWLFCPADRPERFAKAAAA-ADVVILDLEDGVAEAQK
Rv2498c|M.tuberculosis_H37Rv -----MNLRAAGPGWLFCPADRPERFAKAAAA-ADVVILDLEDGVAEAQK
Mflv_2556|M.gilvum_PYR-GCK -----MTL-APGTAWLFCPADRPERFEKAAAA-ADIVILDLEDGVAAKDR
TH_0719|M.thermoresistible__bu -----MDLSAVGPAWLFCPADRPERFEKAAAA-ADVVILDLEDGVAAKDR
MAB_1369|M.abscessus_ATCC_1997 ---MTAPAYRPRRTTLAVPGSSQKMIDKAKTLPADEVFLDLEDAVAAPAK
Mvan_4487|M.vanbaalenii_PYR-1 ----MENRYRPRRTCLSVPGSSAKMIQKAKTLPADEVFLDLEDAVAPGAK
* *. . .* : * : :****.** :
MSMEG_6298|M.smegmatis_MC2_155 DAARDAAVTALSALPTAGVPVLCVRINAVDSGFAEDDLTALEPVLHRLDL
MAV_5074|M.avium_104 ARAREAVDR---WLATGGVG--TVRINGTETPWHQEDL---EMAARHGCP
MMAR_4158|M.marinum_M AQARRDALNFLCTTTPAKTM-AAIRVNAPDTLDGWRDLTALLDSAADPDF
MUL_4021|M.ulcerans_Agy99 AQARRDALNFLCTTTPAKTM-AAIRVNAPDTLDGWRDLTALLDSAADPDF
Mb2526c|M.bovis_AF2122/97 PAAR----NALRDTPLDPER-TVVRINAGGTADQARDLEALAGTAYT--T
Rv2498c|M.tuberculosis_H37Rv PAAR----NALRDTPLDPER-TVVRINAGGTADQARDLEALAGTAYT--T
Mflv_2556|M.gilvum_PYR-GCK QAAR----EALVNTPLDPAR-TVVRVNPTGTADHGPDLEAVARTAYT--T
TH_0719|M.thermoresistible__bu PAAR----EALLSTPLDPAR-TVVRVNPAGTADHRLDLDALTRTPYR--T
MAB_1369|M.abscessus_ATCC_1997 VEARRRIAEALNAPGWSGQLLSVRVNDWTTEWTYADLVEIVSRAGANIDN
Mvan_4487|M.vanbaalenii_PYR-1 EQARGQVAAALAEPGWAGQLRGVRVNDWTTPWTHSDIIEVVARAGASLDI
** : :
MSMEG_6298|M.smegmatis_MC2_155 VMVPMSAEPDRIRRVSELLTRAERSAGADDGRIGIIPLVETAAGILNAPS
MAV_5074|M.avium_104 VMVPKARSASHLAQISRRLPTGT----------ALIALIETAAGVLAAAD
MMAR_4158|M.marinum_M LMLPKVQSAAEIGLVKRLL---------------RQSQKTTRLIATAESA
MUL_4021|M.ulcerans_Agy99 LMLPKVQSAAEIGLVKRLL---------------RQSQKTTRLIATAETA
Mb2526c|M.bovis_AF2122/97 VMLPKAESAAQVIELAPRD---------------VIALVETARGAVCAAE
Rv2498c|M.tuberculosis_H37Rv VMLPKAESAAQVIELAPRD---------------VIALVETARGAVCAAE
Mflv_2556|M.gilvum_PYR-GCK VMLAKTEAPEQVRALAPLD---------------VIVLIETPLGALNVVD
TH_0719|M.thermoresistible__bu VMLAKTDAAEQVRALAPLQ---------------VVVLIETPLGALNVVE
MAB_1369|M.abscessus_ATCC_1997 ILLPKVTAASHVTALDLLLTQLERSHGLQVGAIGIQAQIENAEGLTNIDA
Mvan_4487|M.vanbaalenii_PYR-1 VVLPKVTDASHVHALDLLLSQLEATHGLPLGRIGIEAQIEDAKGLTNVDA
:::. . .: :
MSMEG_6298|M.smegmatis_MC2_155 LASADARVHTLSFGPADLSRELG---------ITPTSDGDEFLFARSQLV
MAV_5074|M.avium_104 ICAVEG-VVRAAFGSVDLGAELG---------IDPE-DHESLRYARSAVV
MMAR_4158|M.marinum_M VVAANPDCALDPRLVDAVIFGSAD----MAADLGAEPSAAVVRHARSTIL
MUL_4021|M.ulcerans_Agy99 VVAANPDCALDPRLVDAVIFGSAD----MAADLGAEPSAAVVRHARSTIL
Mb2526c|M.bovis_AF2122/97 IAAADPTVGMMWG-AEDLIATLGG----SSSRRADGAYRDVARHVRSTIL
Rv2498c|M.tuberculosis_H37Rv IAAADPTVGMMWG-AEDLIATLGG----SSSRRADGAYRDVARHVRSTIL
Mflv_2556|M.gilvum_PYR-GCK LARVDNAYALMWG-AEDLFAELGG----TANRYPDGSYREVARHVRSQTL
TH_0719|M.thermoresistible__bu SARADNAVAVMWG-AEDLFAVTGG----TDNRNPDGSYRDVARHVRSQSL
MAB_1369|M.abscessus_ATCC_1997 IASASPRMRSLVFGPADFMASLGMRTLVVGEQPPGY-DGEAYHHVLMTIL
Mvan_4487|M.vanbaalenii_PYR-1 IASA-PRVQALVLGPADLMASLNMRTLEVGEQPEGYTEGDAYHHVLMRIL
. . .. :
MSMEG_6298|M.smegmatis_MC2_155 LAAAAANCPAPVDGPYLDLDDDDGLIRSARHARKLGFGGKQVIHPSQIAS
MAV_5074|M.avium_104 LGSAAAGLAAPLDGVTTAVHDEGAAGADAARAARLGFGGKLCIHPCQVAP
MMAR_4158|M.marinum_M SHAAAAKIPV-IDAPFFDIDDIPGLRQAVREAVAEGFAGKAAIHPDHVAL
MUL_4021|M.ulcerans_Agy99 SHAAAAKIPV-IDAPFFDIDDIPGLRQAVREAVAEGFAGKAAIHPDHVAL
Mb2526c|M.bovis_AF2122/97 LAASAFGRLA-LDAVHLDILDVEGLQEEARDAAAVGFDVTVCIHPSQIPV
Rv2498c|M.tuberculosis_H37Rv LAASAFGRLA-LDAVHLDILDVEGLQEEARDAAAVGFDVTVCIHPSQIPV
Mflv_2556|M.gilvum_PYR-GCK LAAKAYGRVA-LDSVFLDIKNLDGLRGEVDDAVAVGFDGKVAIHPTQLPV
TH_0719|M.thermoresistible__bu LAAKAHGRLA-LDSVYLDIKDLDGLRAEVDDAVAVGFDAKVAIHPTQVAV
MAB_1369|M.abscessus_ATCC_1997 VAARAHGIDA-IDGPYLKVRDVDAFRTAAARSAALGFDGKWVLHPAQIEA
Mvan_4487|M.vanbaalenii_PYR-1 VAARANGIAA-IDGPYLKVRDVASFRRVAGRSAALGYDGKWVLHPDQIEA
: * . :*. : : . . : *: . :** ::
MSMEG_6298|M.smegmatis_MC2_155 VAGAFVTSDAEIDWARAVDAAFSAAEAEGVSSI--RLADGTFVDYPVAQR
MAV_5074|M.avium_104 VNSQFSPSIEEQAWARKVMAAGSGNGAR--------TVDGCMVDKPVVDR
MMAR_4158|M.marinum_M INSGFLPGQSEINWAREVLKSAQAGAGSVG---------GRMVDEAMARR
MUL_4021|M.ulcerans_Agy99 INSGFLPGQSEINWAREVLKSAQAGPGSVG---------GRMVDEAMARR
Mb2526c|M.bovis_AF2122/97 VRKAYRPSHEKLAWARRVLAASRSERGAFAF-------EGQMVDSPVLTH
Rv2498c|M.tuberculosis_H37Rv VRKAYRPSHEKLAWARRVLAASRSERGAFAF-------EGQMVDSPVLTH
Mflv_2556|M.gilvum_PYR-GCK IRDGYTPTEAQADWARRVLAAAQAERGVFQY-------EGQMVDMPVLRR
TH_0719|M.thermoresistible__bu IREGYTPTPDQVRWARHVLAAARDERGVFQF-------EGIMVDAPVLRR
MAB_1369|M.abscessus_ATCC_1997 GNEIFSPRQDDYDRAELILDAYEWHTSVAGGARGAVMLGDEMIDEASRKM
Mvan_4487|M.vanbaalenii_PYR-1 GNEIFSPRQQAYDHAELILEAYEWHTSQAGGARGAVMLGDEMIDEASRKM
: . *. : : . . ::* .
MSMEG_6298|M.smegmatis_MC2_155 ARAILAGAR----------------
MAV_5074|M.avium_104 AARILARAQGLTSP-----------
MMAR_4158|M.marinum_M ARGILARAQGVNEEEPCMS------
MUL_4021|M.ulcerans_Agy99 ARGILARAQGVNEEEPCMS------
Mb2526c|M.bovis_AF2122/97 AETMLRRAGEATSE-----------
Rv2498c|M.tuberculosis_H37Rv AETMLRRAGEATSE-----------
Mflv_2556|M.gilvum_PYR-GCK AERIVALAP----------------
TH_0719|M.thermoresistible__bu AERIVQLAPDPGR------------
MAB_1369|M.abscessus_ATCC_1997 ALVIAGKGRAAGMARQAAPYEPPTG
Mvan_4487|M.vanbaalenii_PYR-1 ALVIAGKGRAAGMQRTGEGFRPPS-
* : .