For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTATIPVRDEPITFGSAAELSAALHEQDYIADTDLATVVHLATALDRPLLLEGPAGVGKTELAKALAAAG GRRLIRLQCYEGLDDNRVLYEWDYAKQLLHVQMLRDRITAATNGIEDIDEASRYLAGRDFGLYSEAFLSV RPLLDAVLSEEPVVLLIDEVDRTEEAMEALLLEILAERQVTIPEIGTFTARTVPWVVLTSNDTRELSSAL KRRCLHYHLGFPTPQREREIVGVRAPHVEPSVVADVVELARTLRELPLRKSPSISEVVDGARAAALLGTE ASHPLLLSTLVKFTSDREIALRHLGGGLSDNLSGVGAQPVEPSKPAEPATVASRPANTTTAVFRGRGGGR R*
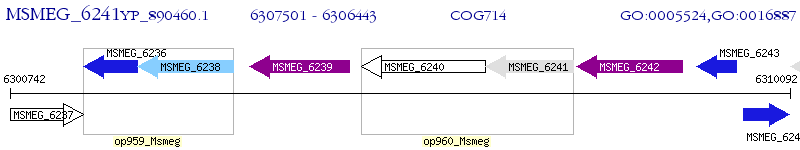
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6241 | - | - | 100% (352) | ATPase associated with various cellular activities, AAA-5 |
| M. smegmatis MC2 155 | MSMEG_4583 | - | 4e-64 | 43.93% (305) | ATPase associated with various cellular activities |
| M. smegmatis MC2 155 | MSMEG_0748 | - | 3e-52 | 42.14% (280) | ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2450c | - | 9e-65 | 45.25% (305) | hypothetical protein Mb2450c |
| M. gilvum PYR-GCK | Mflv_2645 | - | 1e-62 | 46.57% (277) | ATPase |
| M. tuberculosis H37Rv | Rv2426c | - | 3e-65 | 45.25% (305) | hypothetical protein Rv2426c |
| M. leprae Br4923 | MLBr_01810 | moxR | 8e-05 | 33.67% (98) | MoxR homolog |
| M. abscessus ATCC 19977 | MAB_1619 | - | 6e-62 | 45.08% (295) | hypothetical protein MAB_1619 |
| M. marinum M | MMAR_3752 | - | 1e-64 | 45.25% (305) | hypothetical protein MMAR_3752 |
| M. avium 104 | MAV_1748 | - | 5e-64 | 44.92% (305) | ATPase associated with various cellular activities |
| M. thermoresistible (build 8) | TH_0791 | - | 2e-65 | 44.59% (305) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3695 | - | 7e-65 | 45.25% (305) | hypothetical protein MUL_3695 |
| M. vanbaalenii PYR-1 | Mvan_5478 | - | 1e-80 | 54.74% (285) | ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6241|M.smegmatis_MC2_155 MTTATIPVRDE---------PITFGSAAELSAALHEQDYIADTDLATVVH
Mvan_5478|M.vanbaalenii_PYR-1 ----------------------MLDSVEEVREALAGEGYIADERLATTVF
MMAR_3752|M.marinum_M ---MSVPARS----------VPLFADIADVSRRLAQTGYLPDTATATAVF
MUL_3695|M.ulcerans_Agy99 ---MSVPARS----------VPLFADIADVSRRLAQTGYLPDTATATAVF
Mb2450c|M.bovis_AF2122/97 ---MTVPARP----------TPLFADIADVSRRLAETGYLPDTATATAVF
Rv2426c|M.tuberculosis_H37Rv ---MTVPARP----------TPLFADIADVSRRLAETGYLPDTATATAVF
MAV_1748|M.avium_104 ---MSVPARP----------RPLFADIDDVSRRLAETGYLPDTATATAVF
Mflv_2645|M.gilvum_PYR-GCK ---MSVPASI----------QPLFADIDDVARRLAETGYLPDTATATAVF
TH_0791|M.thermoresistible__bu ---VSLPARS----------APLFADIDDVSRRLAETGYLPDTATATAVY
MAB_1619|M.abscessus_ATCC_1997 ---MSDPLRPSGSSQSRTQTAPLFTSIDDVTTRLAETGYLADTATATAVF
MLBr_01810|M.leprae_Br4923 ---MTSAGGLP---------VGADGCSGPGGQSGPRAHEAPPGGADGLVA
. *
MSMEG_6241|M.smegmatis_MC2_155 LATALDRPLLLEGPAGVGKTELAK-ALAAAGGRRLIRLQCYEGLDDNRVL
Mvan_5478|M.vanbaalenii_PYR-1 LQTRLDKPLLIEGPAGVGKTELAK-VLAAATGRRLLRLQCYEGQDETKAL
MMAR_3752|M.marinum_M LADRLGKPLLVEGPAGVGKTELAR-AVAQATGSGLVRLQCYEGVDEARAL
MUL_3695|M.ulcerans_Agy99 LADRLGKPLLVEGPAGVGKTELAR-AVAQATGSGLVRLQCYEGVDEARAL
Mb2450c|M.bovis_AF2122/97 LADRLGKPLLVEGPAGVGKTELAR-AVAQATGSGLVRLQCYEGVDEARAL
Rv2426c|M.tuberculosis_H37Rv LADRLGKPLLVEGPAGVGKTELAR-AVAQATGSGLVRLQCYEGVDEARAL
MAV_1748|M.avium_104 LADRLGKPLLVEGPAGVGKTELAR-AVAQATGSGLVRLQCYEGVDEARAL
Mflv_2645|M.gilvum_PYR-GCK LADRLGKPLLVEGPAGVGKTELAR-AIAQSTGSGLVRLQCYEGVDESRAL
TH_0791|M.thermoresistible__bu LADRLGKPLLVEGPAGVGKTELAR-AVAQATGSELVRLQCYEGVDEARAL
MAB_1619|M.abscessus_ATCC_1997 LADRLGKPLLVEGPAGVGKTELAR-AVAHTTGSGLVRLQCYEGVDEARAL
MLBr_01810|M.leprae_Br4923 EVHTLERAIFEVKRVIVGQDQLVERMLVGLLSKGHVLLEGVPGVAKTLAV
* :.:: . **: :*.. :. . : *: * . .:
MSMEG_6241|M.smegmatis_MC2_155 YEWD--YAKQLLHVQMLRDRITAATNGIEDIDEASRYLAGRDFGLYSEAF
Mvan_5478|M.vanbaalenii_PYR-1 YEWD--YGKQLLYTQILREKIGQLVSDAADLTEAVERIGAQESVFFSDRF
MMAR_3752|M.marinum_M YEWN--HAKQILRIQSGSG----------DWEATKD-------DVFSEEF
MUL_3695|M.ulcerans_Agy99 YEWN--HAKQILRIQSGSG----------DWEATKD-------DVFSEEF
Mb2450c|M.bovis_AF2122/97 YEWN--HAKQILRIQAGSG----------DWEATKT-------DVLSEEF
Rv2426c|M.tuberculosis_H37Rv YEWN--HAKQILRIQAGSG----------DWEATKT-------DVFSEEF
MAV_1748|M.avium_104 YEWN--HAKQILRIQAGSG----------DWDQTKT-------DVFSEEF
Mflv_2645|M.gilvum_PYR-GCK YEWN--HAKQILRIQAGHG----------DWDQTRD-------DVFSEEF
TH_0791|M.thermoresistible__bu YEWN--HAKQILRIQAGNAGA-----GGEAWERTKD-------DVFSEEF
MAB_1619|M.abscessus_ATCC_1997 YEWN--HAKQILRIQAGSG----------DWDETKT-------DVFAEEF
MLBr_01810|M.leprae_Br4923 ETFARVVGGTFARIQFTP-----------DLVPTDIIG----TRIYRQGK
: . : * : . :
MSMEG_6241|M.smegmatis_MC2_155 LSVRPLLDAVLSEEPVVLLIDEVDRTEEAMEALLLEILAERQVTIPEIGT
Mvan_5478|M.vanbaalenii_PYR-1 LAPRPLLEAVRSPEPVVLLVDEIDRADEALEAVLLELLGEYQVSVPEIGT
MMAR_3752|M.marinum_M LLQRPLLTAIRRTEPTVLLIDETDKADIEIEGLLLEVLSDFAVTVPELGT
MUL_3695|M.ulcerans_Agy99 LLQRPLLTAIRRTEPTVLLIDETDKADIEIEGLLLEVLSDFAVTVPELGT
Mb2450c|M.bovis_AF2122/97 LLQRPLLTAIRRTEPTVLLIDETDKADIEIEGLLLEVLSDFAVTVPELGT
Rv2426c|M.tuberculosis_H37Rv LLQRPLLTAIRRTEPTVLLIDETDKADIEIEGLLLEVLSDFAVTVPELGT
MAV_1748|M.avium_104 LLQRPLLTAIRRTEPTVLLIDETDKADIEIEGLLLEVLSDFAVTVPELGT
Mflv_2645|M.gilvum_PYR-GCK LLTRPLLTAIRRTEPTVLLIDETDKADIEIEGLLLEVLSDFAVTVPELGT
TH_0791|M.thermoresistible__bu LLSRPLLTAIRRTEPTVLLIDETDKADIEIEGLLLEVLSDFAVTVPELGT
MAB_1619|M.abscessus_ATCC_1997 LLSRPLLTAIRREDPTVLLIDETDKADIEIEGLLLEVLSDFAVTIPELGT
MLBr_01810|M.leprae_Br4923 EEFDTELGPVVVN---FLLADEINRAPAKVQSALLEVMQERHVSIG--GK
. * .: .** ** ::: ::. ***:: : *:: *.
MSMEG_6241|M.smegmatis_MC2_155 FTARTVPWVVLTSN------DTRELSSALKRRCLH-YHLGFPTPQREREI
Mvan_5478|M.vanbaalenii_PYR-1 FRAVHAPYVVLTSN------NTRDLAAALKRRCLH-LFLDYPAAERELEI
MMAR_3752|M.marinum_M LTAARAPFVLLTSN------ATRELSEALKRRCLF-LHIDFPTPEMERRI
MUL_3695|M.ulcerans_Agy99 LTATRAPFVLLTSN------ATRELSEALKRRCLF-LHIDFPTPEMERRI
Mb2450c|M.bovis_AF2122/97 LTATRAPFVLLTSN------ATRELSEALKRRCLY-LHIDFPTPELERRI
Rv2426c|M.tuberculosis_H37Rv LTATRAPFVLLTSN------ATRELSEALKRRCLY-LHIDFPTPELERRI
MAV_1748|M.avium_104 LTAERPPFVVLTSN------ATRELSEALKRRCLF-LHIDFPSPDLERRI
Mflv_2645|M.gilvum_PYR-GCK ITAERRPLVVLTSN------ATRELSEALKRRCLF-LHIDFPDPDLERRI
TH_0791|M.thermoresistible__bu ITAERIPFVVLTSN------ATRELSEALKRRCLF-LHIDFPDPDLERRI
MAB_1619|M.abscessus_ATCC_1997 IAASRKPFVLLTSN------ATRELSEALKRRCLF-LHIDFPDAELERRI
MLBr_01810|M.leprae_Br4923 TFPLPNPFLVMATQNPIEHEGVYLLPEAQRDRFLFKINVGYPSPEEEREI
. * :::::: . *. * : * *. :.:* .: * .*
MSMEG_6241|M.smegmatis_MC2_155 VGVR--APHVEPSVV--ADVVELARTLRE--LPLRKSPSISEVVDGARAA
Mvan_5478|M.vanbaalenii_PYR-1 VRSK--NTGLSDALA--AQLVDIVRGLRE--LELRKAPSISETIDWARTL
MMAR_3752|M.marinum_M LLSR--VPELPEHIA--EELVRIIGVLRG--MQLKKLPSIAETIDWGRTV
MUL_3695|M.ulcerans_Agy99 LLSR--VPELPEHIA--EELVRIIGVLRG--MQLKKLPSIAETIDWGRTV
Mb2450c|M.bovis_AF2122/97 LLSR--VPELPEHFA--EELVRIIGVLRG--MQLKKVPSIAETIDWGRTV
Rv2426c|M.tuberculosis_H37Rv LLSR--VPELPEHFA--EELVRIIGVLRG--MQLKKVPSIAETIDWGRTV
MAV_1748|M.avium_104 LLSR--VPELPAHLA--DELVRIIGVLRG--MQLKKVPSIAETIDWGRTL
Mflv_2645|M.gilvum_PYR-GCK LLSR--VPELPERLA--EELVRIIGVLRG--MQLKKLPSVAETIDWARTV
TH_0791|M.thermoresistible__bu LLSR--VPELPERIA--SELVRIVGVLRG--MQLKKLPSVAETIDWARTV
MAB_1619|M.abscessus_ATCC_1997 LLKR--VPELPASLA--EELVRIIGVLRN--MQLKKVPSVAETIDWGRTI
MLBr_01810|M.leprae_Br4923 IYRMGVTPPVPKQILNAGDLLRLQEISANNFVHHALVDYVVRVVTATRHP
: . : . ::: : : : ..: *
MSMEG_6241|M.smegmatis_MC2_155 ALLGTEASHPLLLS----TLVKFTSDREIALRHL----------------
Mvan_5478|M.vanbaalenii_PYR-1 AVLGVDELSAQVLSDTVSVVVKYDKDVRKSLDALPRLVDPNATVPEQHSH
MMAR_3752|M.marinum_M LALGLDTIDDAVVAATLGVVLKHQSDQQRATGELR---------------
MUL_3695|M.ulcerans_Agy99 LALGLDTIDDAVVAATLGVVLKHQSDQQRATGELR---------------
Mb2450c|M.bovis_AF2122/97 LALGLDTIDDAVVAATLGVVLKHQSDQQRATGELR---------------
Rv2426c|M.tuberculosis_H37Rv LALGLDTIDDAVVAATLGVVLKHQSDQQRATGELR---------------
MAV_1748|M.avium_104 LALGLDTIDDAVVAATLGVVLKHQSDQQRATGELR---------------
Mflv_2645|M.gilvum_PYR-GCK LALGMDTIDDEVIAATLGVVLKHQSDQIKAAGELR---------------
TH_0791|M.thermoresistible__bu LALGMDTIDDEMIAATLGVVLKHQSDQQRAISELK---------------
MAB_1619|M.abscessus_ATCC_1997 LALGLDTLDDAVIASTLGVVLKHHSDQQRAAGELR---------------
MLBr_01810|M.leprae_Br4923 EQLGMNDVKTWISFGASPRASLGIIKASRSLALVR-----GRDYVIPQDV
** : : . : :
MSMEG_6241|M.smegmatis_MC2_155 ----GGGLSDNLSGVGAQPVEPSKPAEPATVAS------------RPANT
Mvan_5478|M.vanbaalenii_PYR-1 GHGHGHGHDHDHHGPEHHDHDHGDDGRAARAAKDQPGRFTDGYYGTPRRA
MMAR_3752|M.marinum_M -------LN-----------------------------------------
MUL_3695|M.ulcerans_Agy99 -------LN-----------------------------------------
Mb2450c|M.bovis_AF2122/97 -------LN-----------------------------------------
Rv2426c|M.tuberculosis_H37Rv -------LN-----------------------------------------
MAV_1748|M.avium_104 -------LN-----------------------------------------
Mflv_2645|M.gilvum_PYR-GCK -------LN-----------------------------------------
TH_0791|M.thermoresistible__bu -------LN-----------------------------------------
MAB_1619|M.abscessus_ATCC_1997 -------LN-----------------------------------------
MLBr_01810|M.leprae_Br4923 IEVIPDVLRHRLVLTYNALADEISPEIVINRVLQTVALPQVNSFPQQGHS
MSMEG_6241|M.smegmatis_MC2_155 TTAVFRGRGGGRR--
Mvan_5478|M.vanbaalenii_PYR-1 EQAATAGSLGRRRAL
MMAR_3752|M.marinum_M ---------------
MUL_3695|M.ulcerans_Agy99 ---------------
Mb2450c|M.bovis_AF2122/97 ---------------
Rv2426c|M.tuberculosis_H37Rv ---------------
MAV_1748|M.avium_104 ---------------
Mflv_2645|M.gilvum_PYR-GCK ---------------
TH_0791|M.thermoresistible__bu ---------------
MAB_1619|M.abscessus_ATCC_1997 ---------------
MLBr_01810|M.leprae_Br4923 VPSVTQAAAAASGR-