For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSVPASIQPLFADIDDVARRLAETGYLPDTATATAVFLADRLGKPLLVEGPAGVGKTELARAIAQSTGSG LVRLQCYEGVDESRALYEWNHAKQILRIQAGHGDWDQTRDDVFSEEFLLTRPLLTAIRRTEPTVLLIDET DKADIEIEGLLLEVLSDFAVTVPELGTITAERRPLVVLTSNATRELSEALKRRCLFLHIDFPDPDLERRI LLSRVPELPERLAEELVRIIGVLRGMQLKKLPSVAETIDWARTVLALGMDTIDDEVIAATLGVVLKHQSD QIKAAGELRLN
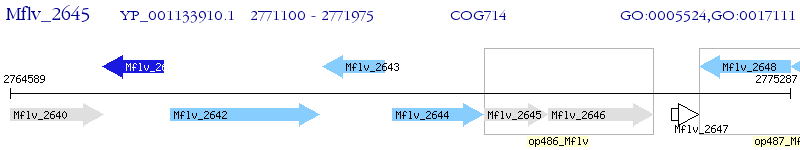
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2645 | - | - | 100% (291) | ATPase |
| M. gilvum PYR-GCK | Mflv_0246 | - | 3e-07 | 28.49% (172) | ATPase |
| M. gilvum PYR-GCK | Mflv_4155 | - | 1e-06 | 24.78% (230) | ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2450c | - | 1e-143 | 87.63% (291) | hypothetical protein Mb2450c |
| M. tuberculosis H37Rv | Rv2426c | - | 1e-144 | 87.97% (291) | hypothetical protein Rv2426c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1619 | - | 1e-138 | 86.22% (283) | hypothetical protein MAB_1619 |
| M. marinum M | MMAR_3752 | - | 1e-145 | 88.32% (291) | hypothetical protein MMAR_3752 |
| M. avium 104 | MAV_1748 | - | 1e-147 | 90.03% (291) | ATPase associated with various cellular activities |
| M. smegmatis MC2 155 | MSMEG_4583 | - | 1e-145 | 88.93% (298) | ATPase associated with various cellular activities |
| M. thermoresistible (build 8) | TH_0791 | - | 1e-145 | 88.51% (296) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3695 | - | 1e-145 | 88.32% (291) | hypothetical protein MUL_3695 |
| M. vanbaalenii PYR-1 | Mvan_3933 | - | 1e-153 | 94.56% (294) | ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2645|M.gilvum_PYR-GCK MSVPASI----------QPLFADIDDVARRLAETGYLPDTATATAVFLAD
Mvan_3933|M.vanbaalenii_PYR-1 MSVPAQT----------VPLFADIDDVARRLAETGYLPDTATATAVFLAD
MSMEG_4583|M.smegmatis_MC2_155 MSVPARP----------APLFADIDDVARKLAETGYLPDTATATAVFLAD
TH_0791|M.thermoresistible__bu VSLPARS----------APLFADIDDVSRRLAETGYLPDTATATAVYLAD
Mb2450c|M.bovis_AF2122/97 MTVPARP----------TPLFADIADVSRRLAETGYLPDTATATAVFLAD
Rv2426c|M.tuberculosis_H37Rv MTVPARP----------TPLFADIADVSRRLAETGYLPDTATATAVFLAD
MMAR_3752|M.marinum_M MSVPARS----------VPLFADIADVSRRLAQTGYLPDTATATAVFLAD
MUL_3695|M.ulcerans_Agy99 MSVPARS----------VPLFADIADVSRRLAQTGYLPDTATATAVFLAD
MAV_1748|M.avium_104 MSVPARP----------RPLFADIDDVSRRLAETGYLPDTATATAVFLAD
MAB_1619|M.abscessus_ATCC_1997 MSDPLRPSGSSQSRTQTAPLFTSIDDVTTRLAETGYLADTATATAVFLAD
:: * ***:.* **: :**:****.********:***
Mflv_2645|M.gilvum_PYR-GCK RLGKPLLVEGPAGVGKTELARAIAQSTGSGLVRLQCYEGVDESRALYEWN
Mvan_3933|M.vanbaalenii_PYR-1 RLGKPLLVEGPAGVGKTELARAIAQSTGSELVRLQCYEGVDESRALYEWN
MSMEG_4583|M.smegmatis_MC2_155 RLGKPLLVEGPAGVGKTELARAVAQCTGSELVRLQCYEGVDEARALYEWN
TH_0791|M.thermoresistible__bu RLGKPLLVEGPAGVGKTELARAVAQATGSELVRLQCYEGVDEARALYEWN
Mb2450c|M.bovis_AF2122/97 RLGKPLLVEGPAGVGKTELARAVAQATGSGLVRLQCYEGVDEARALYEWN
Rv2426c|M.tuberculosis_H37Rv RLGKPLLVEGPAGVGKTELARAVAQATGSGLVRLQCYEGVDEARALYEWN
MMAR_3752|M.marinum_M RLGKPLLVEGPAGVGKTELARAVAQATGSGLVRLQCYEGVDEARALYEWN
MUL_3695|M.ulcerans_Agy99 RLGKPLLVEGPAGVGKTELARAVAQATGSGLVRLQCYEGVDEARALYEWN
MAV_1748|M.avium_104 RLGKPLLVEGPAGVGKTELARAVAQATGSGLVRLQCYEGVDEARALYEWN
MAB_1619|M.abscessus_ATCC_1997 RLGKPLLVEGPAGVGKTELARAVAHTTGSGLVRLQCYEGVDEARALYEWN
**********************:*: *** ************:*******
Mflv_2645|M.gilvum_PYR-GCK HAKQILRIQAG----HG---DWDQTRDDVFSEEFLLTRPLLTAIRRTEPT
Mvan_3933|M.vanbaalenii_PYR-1 HAKQILRIQAG----QGEGADWDRTKDDVFSEEFLLTRPLLTAIRRTEPT
MSMEG_4583|M.smegmatis_MC2_155 HAKQILRIQAGNAAAAGDSDAWEQTKTDVFSEEFLLTRPLLTAIKRTDPT
TH_0791|M.thermoresistible__bu HAKQILRIQAGNAGAGG--EAWERTKDDVFSEEFLLSRPLLTAIRRTEPT
Mb2450c|M.bovis_AF2122/97 HAKQILRIQAG----SG---DWEATKTDVLSEEFLLQRPLLTAIRRTEPT
Rv2426c|M.tuberculosis_H37Rv HAKQILRIQAG----SG---DWEATKTDVFSEEFLLQRPLLTAIRRTEPT
MMAR_3752|M.marinum_M HAKQILRIQSG----SG---DWEATKDDVFSEEFLLQRPLLTAIRRTEPT
MUL_3695|M.ulcerans_Agy99 HAKQILRIQSG----SG---DWEATKDDVFSEEFLLQRPLLTAIRRTEPT
MAV_1748|M.avium_104 HAKQILRIQAG----SG---DWDQTKTDVFSEEFLLQRPLLTAIRRTEPT
MAB_1619|M.abscessus_ATCC_1997 HAKQILRIQAG----SG---DWDETKTDVFAEEFLLSRPLLTAIRREDPT
*********:* * *: *: **::***** *******:* :**
Mflv_2645|M.gilvum_PYR-GCK VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTITAERRPLVVLTSNAT
Mvan_3933|M.vanbaalenii_PYR-1 VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTITAERRPLVVLTSNAT
MSMEG_4583|M.smegmatis_MC2_155 VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTITAERPPFVVLTSNAT
TH_0791|M.thermoresistible__bu VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTITAERIPFVVLTSNAT
Mb2450c|M.bovis_AF2122/97 VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTLTATRAPFVLLTSNAT
Rv2426c|M.tuberculosis_H37Rv VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTLTATRAPFVLLTSNAT
MMAR_3752|M.marinum_M VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTLTAARAPFVLLTSNAT
MUL_3695|M.ulcerans_Agy99 VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTLTATRAPFVLLTSNAT
MAV_1748|M.avium_104 VLLIDETDKADIEIEGLLLEVLSDFAVTVPELGTLTAERPPFVVLTSNAT
MAB_1619|M.abscessus_ATCC_1997 VLLIDETDKADIEIEGLLLEVLSDFAVTIPELGTIAASRKPFVLLTSNAT
****************************:*****::* * *:*:******
Mflv_2645|M.gilvum_PYR-GCK RELSEALKRRCLFLHIDFPDPDLERRILLSRVPELPERLAEELVRIIGVL
Mvan_3933|M.vanbaalenii_PYR-1 RELSEALKRRCLFLHIDFPDPDLERRILLSRVPELPERLAEELVRIIGVL
MSMEG_4583|M.smegmatis_MC2_155 RELSEALKRRCLFLHIDFPDPDLERRILLSRVPELPEHIASELVRIIGVL
TH_0791|M.thermoresistible__bu RELSEALKRRCLFLHIDFPDPDLERRILLSRVPELPERIASELVRIVGVL
Mb2450c|M.bovis_AF2122/97 RELSEALKRRCLYLHIDFPTPELERRILLSRVPELPEHFAEELVRIIGVL
Rv2426c|M.tuberculosis_H37Rv RELSEALKRRCLYLHIDFPTPELERRILLSRVPELPEHFAEELVRIIGVL
MMAR_3752|M.marinum_M RELSEALKRRCLFLHIDFPTPEMERRILLSRVPELPEHIAEELVRIIGVL
MUL_3695|M.ulcerans_Agy99 RELSEALKRRCLFLHIDFPTPEMERRILLSRVPELPEHIAEELVRIIGVL
MAV_1748|M.avium_104 RELSEALKRRCLFLHIDFPSPDLERRILLSRVPELPAHLADELVRIIGVL
MAB_1619|M.abscessus_ATCC_1997 RELSEALKRRCLFLHIDFPDAELERRILLKRVPELPASLAEELVRIIGVL
************:****** .::******.****** :*.*****:***
Mflv_2645|M.gilvum_PYR-GCK RGMQLKKLPSVAETIDWARTVLALGMDTIDDEVIAATLGVVLKHQSDQIK
Mvan_3933|M.vanbaalenii_PYR-1 RGMALKKLPSVAETIDWGRTLLALGMDTVDDEVIAATLGVVLKHQSDQIK
MSMEG_4583|M.smegmatis_MC2_155 RGMQLKKLPSVAETIDWGRTVLALGMDTIDDEMIAATLGVILKHQSDQQR
TH_0791|M.thermoresistible__bu RGMQLKKLPSVAETIDWARTVLALGMDTIDDEMIAATLGVVLKHQSDQQR
Mb2450c|M.bovis_AF2122/97 RGMQLKKVPSIAETIDWGRTVLALGLDTIDDAVVAATLGVVLKHQSDQQR
Rv2426c|M.tuberculosis_H37Rv RGMQLKKVPSIAETIDWGRTVLALGLDTIDDAVVAATLGVVLKHQSDQQR
MMAR_3752|M.marinum_M RGMQLKKLPSIAETIDWGRTVLALGLDTIDDAVVAATLGVVLKHQSDQQR
MUL_3695|M.ulcerans_Agy99 RGMQLKKLPSIAETIDWGRTVLALGLDTIDDAVVAATLGVVLKHQSDQQR
MAV_1748|M.avium_104 RGMQLKKVPSIAETIDWGRTLLALGLDTIDDAVVAATLGVVLKHQSDQQR
MAB_1619|M.abscessus_ATCC_1997 RNMQLKKVPSVAETIDWGRTILALGLDTLDDAVIASTLGVVLKHHSDQQR
*.* ***:**:******.**:****:**:** ::*:****:***:*** :
Mflv_2645|M.gilvum_PYR-GCK AAGELRLN
Mvan_3933|M.vanbaalenii_PYR-1 ASGELRLN
MSMEG_4583|M.smegmatis_MC2_155 AAGELKLN
TH_0791|M.thermoresistible__bu AISELKLN
Mb2450c|M.bovis_AF2122/97 ATGELRLN
Rv2426c|M.tuberculosis_H37Rv ATGELRLN
MMAR_3752|M.marinum_M ATGELRLN
MUL_3695|M.ulcerans_Agy99 ATGELRLN
MAV_1748|M.avium_104 ATGELRLN
MAB_1619|M.abscessus_ATCC_1997 AAGELRLN
* .**:**