For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTPQHLSDRYELGEILGFGGMSEVHLARDLRLHRDVAVKVLRADLARDPSFYLRFRREAQNAAALNHPAI VAVYDTGEAETPNGPLPYIVMEYVDGVTLRDIVHTDGPIAPRRAIEIIADACQALNFSHQHGIIHRDVKP ANIMISKNNAVKVMDFGIARALADTGNSVTQTAAVIGTAQYLSPEQARGETVDARSDVYSLGCVLYEILT GEPPFIGDSPVAVAYQHVREDPVPPSRRHADVTPELDAVVLKALAKNPDNRYQTAAEMRADLIRVHEGQA PDAPKVLTDAERTSMLAAPPADRAGAATQDMPVPRPAGYSKQRSTSVARWLIAVAVLAVLTVVVTVAINM VGGNPRNVQVPDVAEQSADDAQAALQNRGFKTVIDRQPDNEVPPGLVIGTDPEAGSELGAGEQVTINVST GPEQALVPDVAGLTPTQARQKLKDAGFEKFRESPSPSTPEQKGRVLATNPQANQTAAIINEITIVVGAGP EDAPVLSCAGQNAESCKAILAAGGFTNTVVVEVDNPAAAGQVVGTEPADGQSVPKDTVIQIRVSKGNQFV MPDLVGQFWSDAYPRLTALGWTGVLDKGPDVRDSGQRTNAVVTQSPSAGTPVNKDAKITLSFAA*
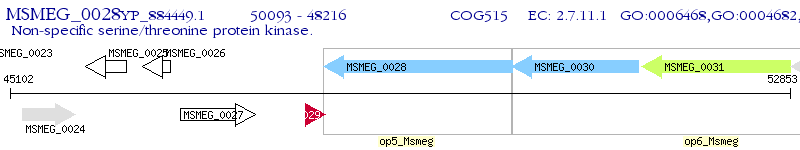
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0028 | - | - | 100% (625) | serine-threonine protein kinase |
| M. smegmatis MC2 155 | MSMEG_0030 | pknA | 1e-60 | 38.61% (360) | serine/threonine protein kinase PknA |
| M. smegmatis MC2 155 | MSMEG_4243 | - | 2e-58 | 38.18% (330) | serine/threonine-protein kinase PK-1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0014c | pknB | 0.0 | 70.03% (624) | transmembrane serine/threonine-protein kinase B PKNB |
| M. gilvum PYR-GCK | Mflv_0812 | - | 0.0 | 74.56% (625) | protein kinase |
| M. tuberculosis H37Rv | Rv0014c | pknB | 0.0 | 70.03% (624) | transmembrane serine/threonine-protein kinase B PKNB |
| M. leprae Br4923 | MLBr_00016 | pknB | 0.0 | 68.20% (629) | putative serine/threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_0033c | - | 0.0 | 64.43% (627) | serine/threonine-protein kinase PknB |
| M. marinum M | MMAR_0016 | pknB | 0.0 | 71.63% (624) | serine/threonine-protein kinase B PknB |
| M. avium 104 | MAV_0017 | - | 0.0 | 71.73% (626) | serine/threonine protein kinase |
| M. thermoresistible (build 8) | TH_2631 | - | 0.0 | 75.96% (628) | PUTATIVE serine/threonine protein kinase |
| M. ulcerans Agy99 | MUL_0018 | pknB | 0.0 | 71.47% (624) | serine/threonine-protein kinase B PknB |
| M. vanbaalenii PYR-1 | Mvan_0023 | - | 0.0 | 76.48% (625) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0812|M.gilvum_PYR-GCK MTTPQHLSDRYEVGEILGFGGMSEVHLARDLRLHRDVAIKVLRADLARDP
Mvan_0023|M.vanbaalenii_PYR-1 MTTPQHLSDRYEVGEILGFGGMSEVHLARDLRLHRDVAIKVLRADLARDP
MSMEG_0028|M.smegmatis_MC2_155 MTTPQHLSDRYELGEILGFGGMSEVHLARDLRLHRDVAVKVLRADLARDP
TH_2631|M.thermoresistible__bu MTTPQHLSDRYELGEILGFGGMSEVHLARDLRLHRDVAVKVLRADLARDP
MLBr_00016|M.leprae_Br4923 MTTPPHLSDRYELGDILGFGGMSEVHLARDIRLHRDVAVKVLRADLARDP
MAV_0017|M.avium_104 MTTPQHLSDRYELGEILGFGGMSEVHLARDVRLHRDVAVKVLRADLARDP
Mb0014c|M.bovis_AF2122/97 MTTPSHLSDRYELGEILGFGGMSEVHLARDLRLHRDVAVKVLRADLARDP
Rv0014c|M.tuberculosis_H37Rv MTTPSHLSDRYELGEILGFGGMSEVHLARDLRLHRDVAVKVLRADLARDP
MMAR_0016|M.marinum_M MTTPQHLSDRYELGDILGFGGMSEVHLARDLRLHRDVAVKVLRADLARDP
MUL_0018|M.ulcerans_Agy99 MTTPQHLSDRYELGDILGFGGMSEVHLARDLRLHRDVAVKVLRADLARDP
MAB_0033c|M.abscessus_ATCC_199 MTTPQHLSDRYELGEILGYGGMSEVHLARDLRLNRDVAIKVLRADLARDP
**** *******:*:***:***********:**:****:***********
Mflv_0812|M.gilvum_PYR-GCK SFYLRFRREAQNAAALNHPAIVAVYDTGEAETPTGPLPYIVMEYVDGVTL
Mvan_0023|M.vanbaalenii_PYR-1 SFYLRFRREAQNAAALNHPAIVAVYDTGEAETPTGPLPYIVMEYVDGVTL
MSMEG_0028|M.smegmatis_MC2_155 SFYLRFRREAQNAAALNHPAIVAVYDTGEAETPNGPLPYIVMEYVDGVTL
TH_2631|M.thermoresistible__bu SFYLRFRREAQNAAALNHPAIVAVYDTGEAETPTGPLPYIVMEYVDGVTL
MLBr_00016|M.leprae_Br4923 SFYLRFRREAQNAAALNHPSIVAVYDTGEAETSAGPLPYIVMEYVDGATL
MAV_0017|M.avium_104 SFYLRFRREAQNAAALNHPSIVAVYDTGEAETPSGPLPYIVMEYVDGVTL
Mb0014c|M.bovis_AF2122/97 SFYLRFRREAQNAAALNHPAIVAVYDTGEAETPAGPLPYIVMEYVDGVTL
Rv0014c|M.tuberculosis_H37Rv SFYLRFRREAQNAAALNHPAIVAVYDTGEAETPAGPLPYIVMEYVDGVTL
MMAR_0016|M.marinum_M SFYLRFRREAQNAAALNHPAIVAVYDTGEAETPSGPLPYIVMEYVNGVTL
MUL_0018|M.ulcerans_Agy99 SFYLRFRREAQNAAALNHPAIVAVYDTGEAETPSGPLPYIVMEYVNGFTL
MAB_0033c|M.abscessus_ATCC_199 SFYLRFRREAQNAAALNHPAIVAVYDTGEAETATGPLPYIVMEYVDGITL
*******************:************. ***********:* **
Mflv_0812|M.gilvum_PYR-GCK RDIVHSDGPMPPQRAIEVIADACQALNFSHQHGIIHRDVKPANIMISKTG
Mvan_0023|M.vanbaalenii_PYR-1 RDIVHNDGPMAPQRAIEVIADACQALNFSHQHGIIHRDVKPANIMISKTG
MSMEG_0028|M.smegmatis_MC2_155 RDIVHTDGPIAPRRAIEIIADACQALNFSHQHGIIHRDVKPANIMISKNN
TH_2631|M.thermoresistible__bu RDIVHTDGPMEPKRAIEVIADACQALNFSHQHGIIHRDVKPANIMISKTG
MLBr_00016|M.leprae_Br4923 RDIVHTDGPMPPQQAIEIVADACQALNFSHQNGIIHRDVKPANIMISATN
MAV_0017|M.avium_104 RDIVHTDGPLPPRRAIEIIADACQALNFSHQNGIIHRDVKPANIMISTTN
Mb0014c|M.bovis_AF2122/97 RDIVHTEGPMTPKRAIEVIADACQALNFSHQNGIIHRDVKPANIMISATN
Rv0014c|M.tuberculosis_H37Rv RDIVHTEGPMTPKRAIEVIADACQALNFSHQNGIIHRDVKPANIMISATN
MMAR_0016|M.marinum_M RDIVHTDGPMAPTRAIEIIADACQALNFSHQNGIIHRDVKPANIMISTTN
MUL_0018|M.ulcerans_Agy99 RDIVHTDGPMAPTRAIEIIADACQALNFSHQNGIIHRDVKPANIMISTTN
MAB_0033c|M.abscessus_ATCC_199 RDIVKDDGPMPIRRAIEVIADSCQALNFSHQHGIIHRDVKPANIMISKTG
****: :**: :***::**:*********:*************** ..
Mflv_0812|M.gilvum_PYR-GCK AVKVMDFGIARALADAGNPVTQTAAVIGTAQYLSPEQARGVKVDARSDVY
Mvan_0023|M.vanbaalenii_PYR-1 AVKVMDFGIARALADAGNPVTQTAAVIGTAQYLSPEQARGVKVDARSDVY
MSMEG_0028|M.smegmatis_MC2_155 AVKVMDFGIARALADTGNSVTQTAAVIGTAQYLSPEQARGETVDARSDVY
TH_2631|M.thermoresistible__bu AVKVMDFGIARAIADS-NSVTQTAAVIGTAQYLSPEQARGETVDARSDVY
MLBr_00016|M.leprae_Br4923 AVKVMDFGIARAIADS-TSVTQTAAVIGTAQYLSPEQARGDSVDARSDVY
MAV_0017|M.avium_104 AVKVMDFGIARAIADSGNSVTQTAAVIGTAQYLSPEQARGDAVDARSDVY
Mb0014c|M.bovis_AF2122/97 AVKVMDFGIARAIADSGNSVTQTAAVIGTAQYLSPEQARGDSVDARSDVY
Rv0014c|M.tuberculosis_H37Rv AVKVMDFGIARAIADSGNSVTQTAAVIGTAQYLSPEQARGDSVDARSDVY
MMAR_0016|M.marinum_M AVKVMDFGIARAIADSGNSVTQTAAVIGTAQYLSPEQARGDSVDARSDVY
MUL_0018|M.ulcerans_Agy99 AVKVMDFGIARAIADSGNSVTQTAAVIGTAQYLSPEQARGDSVDARSDVY
MAB_0033c|M.abscessus_ATCC_199 AVKVMDFGIARAVSDAGNSVTQTAAVIGTAQYLSPEQARGETVDARSDVY
************::*: ..********************* ********
Mflv_0812|M.gilvum_PYR-GCK SLGCVLYEMLTGEPPFVGDSPVAVAYQHVREDPVPPSQRHTGISPELDAV
Mvan_0023|M.vanbaalenii_PYR-1 SLGCVLYEVLTGEPPFVGDSPVAVAYQHVREDPVPPSQRHAGISPELDAV
MSMEG_0028|M.smegmatis_MC2_155 SLGCVLYEILTGEPPFIGDSPVAVAYQHVREDPVPPSRRHADVTPELDAV
TH_2631|M.thermoresistible__bu SLGCVLYEILTGEPPFVGDSPVAVAYQHVREDPVPPSRRNPAVSPELDAV
MLBr_00016|M.leprae_Br4923 SLGCVLYEILTGEPPFIGDSPVSVAYQHVREDPIPPSQRHEGISVDLDAV
MAV_0017|M.avium_104 SLGCVLYEILTGEPPFTGDSPVAVAYQHVREDPVPPSQRHEGISADLDAV
Mb0014c|M.bovis_AF2122/97 SLGCVLYEVLTGEPPFTGDSPVSVAYQHVREDPIPPSARHEGLSADLDAV
Rv0014c|M.tuberculosis_H37Rv SLGCVLYEVLTGEPPFTGDSPVSVAYQHVREDPIPPSARHEGLSADLDAV
MMAR_0016|M.marinum_M SLGCVLYEILTGEPPFTGDSPVAVAYQHVREDPIPPSERHEGISAELDAV
MUL_0018|M.ulcerans_Agy99 SLGCVLYEILTGEPPFTGDSPVAVAYQHVREDPIPPSERHEGIFAELDAV
MAB_0033c|M.abscessus_ATCC_199 SLGCVLYELVTGEPPFVGDSPVAVAYQHVREDPVAPSKHNPEIPPGLDSV
********::****** *****:**********:.** :: : **:*
Mflv_0812|M.gilvum_PYR-GCK VLKALAKNPDNRYQTAAEMRSDLVKVHSGEIPDAPKVFTDAERTSLLAST
Mvan_0023|M.vanbaalenii_PYR-1 VLKALAKNPDNRYQTAAEMRTDLVRVHSGEAPDAPKVFTDAERTSLLAAN
MSMEG_0028|M.smegmatis_MC2_155 VLKALAKNPDNRYQTAAEMRADLIRVHEGQAPDAPKVLTDAERTSMLAAP
TH_2631|M.thermoresistible__bu VLKALAKNPDNRYQTAAEMRADLVRVHSGEQPEAPKVLTDAERTSLLASG
MLBr_00016|M.leprae_Br4923 VLKALAKNPENRYQTAAEMRADLIRVHSGQPPEAPKVLTDADRSCLLSSG
MAV_0017|M.avium_104 VLKALAKNPDNRYQTAAEMRADLVRVHNGEKPEAPKVLTDAERSSLLSSG
Mb0014c|M.bovis_AF2122/97 VLKALAKNPENRYQTAAEMRADLVRVHNGEPPEAPKVLTDAERTSLLSSA
Rv0014c|M.tuberculosis_H37Rv VLKALAKNPENRYQTAAEMRADLVRVHNGEPPEAPKVLTDAERTSLLSSA
MMAR_0016|M.marinum_M VLKALAKNPDNRYQTAAEMRADLVRVHNGESPEAPKVLTGAERRSLTASS
MUL_0018|M.ulcerans_Agy99 VLKALAKNPDNRYQTAAEMRADLVRVHNGEAPEAPKVLTGAERRSLTTSS
MAB_0033c|M.abscessus_ATCC_199 VLKALSKNPDNRYQNAAEMRADLMRVYQGEQPEAPKVLTDAERSTMLSTP
*****:***:****.*****:**::*:.*: *:****:*.*:* : ::
Mflv_0812|M.gilvum_PYR-GCK PAHLRTEPVDPVG-PHQPRYDERERGGSLGRWLIAVAVLAVLTVLVTIGI
Mvan_0023|M.vanbaalenii_PYR-1 PAHHRTEPVEPAG-RRQPRYAEPERGGSLGRWLIAVAILAVLTVVVTIGI
MSMEG_0028|M.smegmatis_MC2_155 PADRAGAATQDMP-VPRPAGYSKQRSTSVARWLIAVAVLAVLTVVVTVAI
TH_2631|M.thermoresistible__bu PAHDRTQPTDAVIDPVIPRYTGSDRGGSVGRWVAAVAVLAVLTVVVTVAI
MLBr_00016|M.leprae_Br4923 AGNFGVPRTDALSRQSLDETESD---GSIGRWVAVVAVLAVLTIAIVAAF
MAV_0017|M.avium_104 AGAAGPSRTDPLPRQVLADSGEDRTVGSVGRWIVAVAALAVLTIIVVIAF
Mb0014c|M.bovis_AF2122/97 AGNLSGPRTDPLPRQDLDDTDRDRSIGSVGRWVAVVAVLAVLTVVVTIAI
Rv0014c|M.tuberculosis_H37Rv AGNLSGPRTDPLPRQDLDDTDRDRSIGSVGRWVAVVAVLAVLTVVVTIAI
MMAR_0016|M.marinum_M PSSPAAPRTDPLPRQTLDDTDRDRSTPSVGRWVAVVAVLAVLTIVVTIAI
MUL_0018|M.ulcerans_Agy99 PSSPAAPRTDPLPRQTLDDTDRDRSTPSVGRWVAVVAVLAVLTIVVTIAI
MAB_0033c|M.abscessus_ATCC_199 ANTRAFPASGPQT-GPLPSQYEEPRRNPLSRWLVGLAALVVLTIIVTFAI
. .:.**: :* *.***: :. .:
Mflv_0812|M.gilvum_PYR-GCK NAFGGETREVQVPDVSGQASADAIATLQNRGFSIRTQQKPDSEVPPDHVI
Mvan_0023|M.vanbaalenii_PYR-1 NMFGGGTRDVQVPDVRGQASADAIATLQNRGFSIRTQQKPDSEVPPDHVI
MSMEG_0028|M.smegmatis_MC2_155 NMVGGNPRNVQVPDVAEQSADDAQAALQNRGFKTVIDRQPDNEVPPGLVI
TH_2631|M.thermoresistible__bu NMIGGNPRNVQVPDVSGQASADAIAALQNRGFKTRTQQQPDNSVPPDRVI
MLBr_00016|M.leprae_Br4923 NTFGGNTRDVQVPDVRGQVSADAISALQNRGFKTRTLQKPDSTIPPDHVI
MAV_0017|M.avium_104 NTFGGGARDVQVPDVRGQVSADAIAALQNRGFKTRTLQKPDSAIPPDHVI
Mb0014c|M.bovis_AF2122/97 NTFGGITRDVQVPDVRGQSSADAIATLQNRGFKIRTLQKPDSTIPPDHVI
Rv0014c|M.tuberculosis_H37Rv NTFGGITRDVQVPDVRGQSSADAIATLQNRGFKIRTLQKPDSTIPPDHVI
MMAR_0016|M.marinum_M NTFGGNTRNVQVPNVRGQASADAIAALQNRGFKTRTLLKPDSTIPPDHVI
MUL_0018|M.ulcerans_Agy99 NTFGGNTRNVQVPNVRGQASADAIAALQNRGFKTRTLLKPDSTIPPDHVI
MAB_0033c|M.abscessus_ATCC_199 ILGNGPSRDVQVPDVTGKSQSDAVAALQNLGFKTKVQKNTSSKVAPEKVI
.* .*:****:* : ** ::*** **. :... :.* **
Mflv_0812|M.gilvum_PYR-GCK DTDPNANTSVAAGDEITLNVSTGPEQREVPDVSGLSYADAVRRLTDAGFE
Mvan_0023|M.vanbaalenii_PYR-1 DTDPGANTEVGAGDEITLNVSTGPEQREVPDVSGLSYTEAVRRLTEAGFE
MSMEG_0028|M.smegmatis_MC2_155 GTDPEAGSELGAGEQVTINVSTGPEQALVPDVAGLTPTQARQKLKDAGFE
TH_2631|M.thermoresistible__bu STDPPADTSVSAGDEITVNVSTGPQQREIPDVSSLTYAEAVRRLSAAGFE
MLBr_00016|M.leprae_Br4923 STEPGANASVGAGDEITINVSTGPEQREVPDVSSLNYTDAVKKLTSSGFK
MAV_0017|M.avium_104 GTDPGANASVSAGDEITINVSTGPEQREVPDVSSLSYSDAVTKLKAAGFS
Mb0014c|M.bovis_AF2122/97 GTDPAANTSVSAGDEITVNVSTGPEQREIPDVSTLTYAEAVKKLTAAGFG
Rv0014c|M.tuberculosis_H37Rv GTDPAANTSVSAGDEITVNVSTGPEQREIPDVSTLTYAEAVKKLTAAGFG
MMAR_0016|M.marinum_M GTDPAANASVSAGDQITINVSTGPEQRELPDVSSMTYAEAVKKLTAAGFG
MUL_0018|M.ulcerans_Agy99 GTDPAANASVSAGDQITINVSTGPEQRELPDVSSMTYAEAVKKLTAAGFG
MAB_0033c|M.abscessus_ATCC_199 DTDPAKGSTVSAGAEVTVNVSIGPENQQVPDCHGLSFGDAVKKLQAAGFK
.*:* .: :.** ::*:*** **:: :** :. :* :* :**
Mflv_0812|M.gilvum_PYR-GCK -RFRQTTSTSLPEQKDRVLSTVPPANQTSAITNEITVVIGKGPATAVVPE
Mvan_0023|M.vanbaalenii_PYR-1 -KFRQTASTSLPEQKDRVLSTVPPANQTSAITNEITIVVGKGPATAVVPE
MSMEG_0028|M.smegmatis_MC2_155 -KFRESPSPSTPEQKGRVLATNPQANQTAAIINEITIVVGAGPEDAPVLS
TH_2631|M.thermoresistible__bu -KFRQSESPSTPELRGKVVATNPPANQTAAITNEITVVVGTGPATREVPD
MLBr_00016|M.leprae_Br4923 -SFKQANSPSTPELLGKVIGTNPSANQTSAITNVITIIVGSGPETKQIPD
MAV_0017|M.avium_104 -KFKQANSPSTPELLGKVIGTNPPANQTSAITNVITLIVGSGPETKQVPD
Mb0014c|M.bovis_AF2122/97 -RFKQANSPSTPELVGKVIGTNPPANQTSAITNVVIIIVGSGPATKDIPD
Rv0014c|M.tuberculosis_H37Rv -RFKQANSPSTPELVGKVIGTNPPANQTSAITNVVIIIVGSGPATKDIPD
MMAR_0016|M.marinum_M -RFKEAKSPSTPELMGKVIGTNPPANQTSAITNVITIIVGSGPATKQIPD
MUL_0018|M.ulcerans_Agy99 -RFKEAKSPSTPELMGKVIGTNPPANQTSAITNVITIIVGSGPATKQIPD
MAB_0033c|M.abscessus_ATCC_199 GTFTQSETKSLPEDKGRVMATNPPAGQYSAVTNNITIVVGKGPGEKPVPD
* :: : * ** .:*:.* * *.* :*: * : :::* ** : .
Mflv_0812|M.gilvum_PYR-GCK CVGQTVETCQQILSASGFTK-AVPVEVDSTVSAGQVVGIEPAPGQTVPQD
Mvan_0023|M.vanbaalenii_PYR-1 CVGQTVEVCQQILSASGFTK-SVPVEVDSTVAAGQVVGLEPAAGQTVPQD
MSMEG_0028|M.smegmatis_MC2_155 CAGQNAESCKAILAAGGFTN-TVVVEVDNPAAAGQVVGTEPADGQSVPKD
TH_2631|M.thermoresistible__bu ITGQTVELAERNLSVYGFTK-FTQSAVDSTAPAGQVVGTNPPAGTTVSLD
MLBr_00016|M.leprae_Br4923 VTGQIVEIAQKNLNVYGFTK-FSQASVDSPRPTGEVIGTNPPKDATVPVD
MAV_0017|M.avium_104 IAGQTVDIAQKNLTVYGFTK-ITQVQVDSPRPAGEVIGTNPPKGQTVPVD
Mb0014c|M.bovis_AF2122/97 VAGQTVDVAQKNLNVYGFTK-FSQASVDSPRPAGEVTGTNPPAGTTVPVD
Rv0014c|M.tuberculosis_H37Rv VAGQTVDVAQKNLNVYGFTK-FSQASVDSPRPAGEVTGTNPPAGTTVPVD
MMAR_0016|M.marinum_M VAGQSVEEAEQNLNVYGFTK-FSQASVDSPKPAGSVIATNPPAGTTVPID
MUL_0018|M.ulcerans_Agy99 VAGQSVEEAQQNLNVYGFTK-FSQASVDSPKPAGSVIATNPPAGTTVPID
MAB_0033c|M.abscessus_ATCC_199 LGGQNIDVAIKNLPTYGFTAPPTVIPVPSPESKGQILSTNPPANTMFPED
** : . * . *** * .. . *.: . :*. . .. *
Mflv_0812|M.gilvum_PYR-GCK SVIQIQVSRGNQFVMPDLTGQFWTDAEPRLRALGWTGVLDKGGDVQNSGQ
Mvan_0023|M.vanbaalenii_PYR-1 TVIQIQVSRGNQFVMPDLRGQFWADAEPRLRALGWTGVLDKGADVPNSGQ
MSMEG_0028|M.smegmatis_MC2_155 TVIQIRVSKGNQFVMPDLVGQFWSDAYPRLTALGWTGVLDKGPDVRDSGQ
TH_2631|M.thermoresistible__bu SVIELEVSKGNQFVMPDLIGMFWTDAEPRLRALGWTGVLDKGADIPNSGQ
MLBr_00016|M.leprae_Br4923 SVIELQVSKGNQFVMPDLSGMFWADAEPRLRALGWTGVLDKGPDVDAGGS
MAV_0017|M.avium_104 SVIELQVSKGNQFIMPDLSGMFWTDAEPRLRALGWTGILDKGPDVDAGGA
Mb0014c|M.bovis_AF2122/97 SVIELQVSKGNQFVMPDLSGMFWVDAEPRLRALGWTGMLDKGADVDAGGS
Rv0014c|M.tuberculosis_H37Rv SVIELQVSKGNQFVMPDLSGMFWVDAEPRLRALGWTGMLDKGADVDAGGS
MMAR_0016|M.marinum_M SVIELQVSKGNQFVMPDLSGMFWTDAEPRLRALGWTGSLDKGPDVDAGGS
MUL_0018|M.ulcerans_Agy99 SVIELQVSKGNQFVMPDLSGMFWTDAEPRLRALGWTGSLDKGPDVDGGGS
MAB_0033c|M.abscessus_ATCC_199 GVIEIRVSNGGQFRMPNIMGKFWTDAEPMLYQAGWQGGGLSTEDVPSDGN
**::.**.*.** **:: * ** ** * * ** * . *: .*
Mflv_0812|M.gilvum_PYR-GCK RTNAVVRQSPSAGSGVNYGATITLNFAS-
Mvan_0023|M.vanbaalenii_PYR-1 RTNAVVTQSPSPGSGVNYGSRITLNFAS-
MSMEG_0028|M.smegmatis_MC2_155 RTNAVVTQSPSAGTPVNKDAKITLSFAA-
TH_2631|M.thermoresistible__bu RANAVVRQNPSPGAGVNYDARITLNFAS-
MLBr_00016|M.leprae_Br4923 QHNRVAYQNPPAGAGVNRDGIITLKFGQ-
MAV_0017|M.avium_104 QSHRVVYQNPPAGAGVNRDGIITLKFGQ-
Mb0014c|M.bovis_AF2122/97 QHNRVVYQNPPAGTGVNRDGIITLRFGQ-
Rv0014c|M.tuberculosis_H37Rv QHNRVVYQNPPAGTGVNRDGIITLRFGQ-
MMAR_0016|M.marinum_M QHNRVVYQNPPAGAGVNRDGIVTLKFGQ-
MUL_0018|M.ulcerans_Agy99 QHNRVVYQNPPAGAGVNRDGIVTLKFGQ-
MAB_0033c|M.abscessus_ATCC_199 NRARVVFQDPPAGAGVGYNGPIKLRFGAG
. *. *.*..*: *. .. :.* *.