For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVDVSAQEETGRGAQGVLQVGPLKPSLNQTLVSTYGAYVLPDGPGRAAFLAEHGDRVTVAVTSGRTGVDA ELMSALPNLGAVVNFGVGYDTTDVDAAAARDIVVSNTPDVLSDCVADTAVGLLIDVMRKFSASDRYVRAR RWVTEGNYPLAHKVSGSRVGIIGLGRIGTAIATRLGAFGCTISYHNRREIEGSGYRYVGSAADLAAQVDV LIIAAAGGAGTRHLVDRAVLDALGPDGYLVNIARGSVVDEDALVEALADGRLAGAGLDVFTDEPNVPEAL LGMENVVLLPHVGSATVETRNAMEALTLANLDAYLKTGELVTPVPMPAAAPAPTPVQ
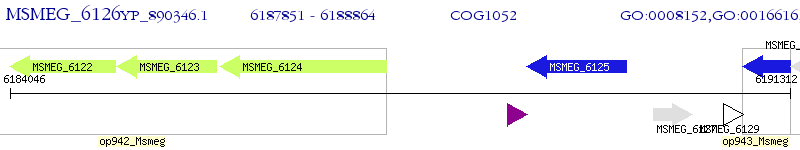
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6126 | - | - | 100% (337) | D-isomer specific 2-hydroxyacid dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6299 | - | 1e-48 | 40.41% (292) | glyoxylate reductase |
| M. smegmatis MC2 155 | MSMEG_2529 | - | 1e-42 | 38.89% (288) | glyoxylate reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 5e-36 | 35.61% (264) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5240 | - | 1e-112 | 67.96% (309) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 5e-36 | 35.61% (264) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 2e-36 | 34.14% (290) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3304c | - | 9e-34 | 34.15% (284) | D-3-phosphoglycerate dehydrogenase |
| M. marinum M | MMAR_1715 | serA1 | 4e-34 | 34.02% (291) | D-3-phosphoglycerate dehydrogenase SerA1 |
| M. avium 104 | MAV_0123 | - | 1e-103 | 61.22% (312) | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_2037 | serA | 1e-35 | 34.85% (264) | PROBABLE D-3-PHOSPHOGLYCERATE DEHYDROGENASE SERA1 (PGDH) |
| M. ulcerans Agy99 | MUL_1952 | serA1 | 5e-34 | 34.02% (291) | D-3-phosphoglycerate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1025 | - | 1e-121 | 70.98% (317) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1715|M.marinum_M ------------MNLPVVLIADKLAQSTVAALGDQVEVRWVDGPDR---E
MUL_1952|M.ulcerans_Agy99 ------------MNLPVVLIADKLAQSTVAALGDQVEVRWVDGPDR---E
Mb3020c|M.bovis_AF2122/97 ------------MSLPVVLIADKLAPSTVAALGDQVEVRWVDGPDR---D
Rv2996c|M.tuberculosis_H37Rv ------------MSLPVVLIADKLAPSTVAALGDQVEVRWVDGPDR---D
MLBr_01692|M.leprae_Br4923 ------------MDLPVVLIADKLAQSTVAALGDQVEVRWVDGPDR---T
MAB_3304c|M.abscessus_ATCC_199 -----------------MLIADKLAPSTVEALGDGVEVRWVDGPDR---P
TH_2037|M.thermoresistible__bu ------------VSLPVVLIADKLAESTVAALGDQVEVRWVDGPDR---E
Mflv_5240|M.gilvum_PYR-GCK ---MSSEGLPECNVGGSVLQVGPLMPSLVRKLADDYDARRLPSDPDERAA
Mvan_1025|M.vanbaalenii_PYR-1 ---MGTE------ENRSVLQVGPLMPSLSRRLADDYAARRLPTEPTERAA
MSMEG_6126|M.smegmatis_MC2_155 MVDVSAQEETG-RGAQGVLQVGPLKPSLNQTLVSTYGAYVLPDGPG-RAA
MAV_0123|M.avium_104 ---MTKLS--------GVFRVGELEPAFGEELAARYDVPRLPDGAA-RDR
:: .. * : * . :
MMAR_1715|M.marinum_M KLLAAVPEADALLVRSATTVDAEVLAAATKLKIVARAGVGLDNVDVDAAT
MUL_1952|M.ulcerans_Agy99 KLLAAVPEADALLVRSATTVDAEVLAAATKLKIVARAGVGLDNVDVDAAT
Mb3020c|M.bovis_AF2122/97 KLLAAVPEADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAAT
Rv2996c|M.tuberculosis_H37Rv KLLAAVPEADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAAT
MLBr_01692|M.leprae_Br4923 KLLAAVPEADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAAT
MAB_3304c|M.abscessus_ATCC_199 ALLAAVPEADALLVRSATTVDAEVLAAGTKLKIVARAGVGLDNVDVKAAT
TH_2037|M.thermoresistible__bu KLLAAVADADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVEAAT
Mflv_5240|M.gilvum_PYR-GCK FLADHGAEFTVAVTSGGVGVDASLMQALPNLGAVVNFGVGYDTTDVEAAH
Mvan_1025|M.vanbaalenii_PYR-1 FLAERGAEIIAVVTSGRTGVDAALMDALPNLAAVINFGVGYDTTDVAAAA
MSMEG_6126|M.smegmatis_MC2_155 FLAEHGDRVTVAVTSGRTGVDAELMSALPNLGAVVNFGVGYDTTDVDAAA
MAV_0123|M.avium_104 FLAEHAAEVRALLTWGRPGVDAALIAALPNLEVIVNNGAGVDLIDTAAAD
* . :. . *** :: * .:* : . *.* * *. **
MMAR_1715|M.marinum_M TRGVLVVNAPTSNIHSAAEHAMALLLAAARQIPAADASLREHTWKRSSFS
MUL_1952|M.ulcerans_Agy99 TRGVLVVNAPTSNIHSAAEHAMALLLAAARQIPAADASLREHTWKRSSFS
Mb3020c|M.bovis_AF2122/97 ARGVLVVNAPTSNIHSAAEHALALLLAASRQIPAADASLREHTWKRSSFS
Rv2996c|M.tuberculosis_H37Rv ARGVLVVNAPTSNIHSAAEHALALLLAASRQIPAADASLREHTWKRSSFS
MLBr_01692|M.leprae_Br4923 ARGVLVVNAPTSNIHSAAEHALALLLAASRQIAEADASLRAHIWKRSSFS
MAB_3304c|M.abscessus_ATCC_199 ARGVLVVNAPTSNIHSAAEHAVTLLLATARQIPAADASLKAHTWKRSSFN
TH_2037|M.thermoresistible__bu ARGVLVVNAPTSNIHSAAEHAIALLLAAARQIPAADATLRQRTWKRSEFS
Mflv_5240|M.gilvum_PYR-GCK ARGIGVSNTPDVLTDCTADTAVGLLIDTMRQLPAADRYVRAGRWPVDGMF
Mvan_1025|M.vanbaalenii_PYR-1 ARGIGVSNTPDVLTDCVADTAVGLMIDTLRQFSASDRYVRAGRWPVDGMY
MSMEG_6126|M.smegmatis_MC2_155 ARDIVVSNTPDVLSDCVADTAVGLLIDVMRKFSASDRYVRARRWVTEGNY
MAV_0123|M.avium_104 RRGIGVSNTPDVLSDTVADTALGLILMTLRRLGAADRYVRAGRWAREGPF
*.: * *:* . .*: *: *:: . *:: :* :: * .
MMAR_1715|M.marinum_M --GAEIFGKTVGVVGLGRIGQLVAQRVEAFGAHVVAYDPYVSPARAAQLG
MUL_1952|M.ulcerans_Agy99 --GAEIFGKTVGVVGLGRIGQLVAQRVEAFGAHVVAYDPYVSPARAAQLG
Mb3020c|M.bovis_AF2122/97 --GTEIFGKTVGVVGLGRIGQLVAQRIAAFGAYVVAYDPYVSPARAAQLG
Rv2996c|M.tuberculosis_H37Rv --GTEIFGKTVGVVGLGRIGQLVAQRIAAFGAYVVAYDPYVSPARAAQLG
MLBr_01692|M.leprae_Br4923 --GTEIFGKTVGVVGLGRIGQLVAARIAAFGAHVIAYDPYVAPARAAQLG
MAB_3304c|M.abscessus_ATCC_199 --GTEIFGKTVGVVGLGRIGQLFAQRLAAFGTHIVAYDPYVSAARAAQLG
TH_2037|M.thermoresistible__bu --GVEIYDKTVGVVGLGRIGQLVAQRLAAFGTHLIAYDPYVSPARAAQLG
Mflv_5240|M.gilvum_PYR-GCK PLTRDVSNSTVGIIGLGRIGTAIAQRLKAFRCSIAYHNRHRVTDCPYPY-
Mvan_1025|M.vanbaalenii_PYR-1 PLTRQVSKTNVGIIGLGRIGAAIALRLKAFGCTISYHNRHEVPDSPYTY-
MSMEG_6126|M.smegmatis_MC2_155 PLAHKVSGSRVGIIGLGRIGTAIATRLGAFGCTISYHNRREIEGSGYRY-
MAV_0123|M.avium_104 PYGRDVSGLQVGILGLGRIGSAIATRLRGFDCAIAYHNRRRIDGSPYRY-
.: **::****** .* *: .* : ::
MMAR_1715|M.marinum_M IELLPLDDLLARADFISVHLPKTPETAGLLGKEALAKTKPGVIIVNAARG
MUL_1952|M.ulcerans_Agy99 IELLPLDDLLARADFISVHLPKTPETAGLLGKEALAKTKPGVIIVNAARG
Mb3020c|M.bovis_AF2122/97 IELLSLDDLLARADFISVHLPKTPETAGLIDKEALAKTKPGVIIVNAARG
Rv2996c|M.tuberculosis_H37Rv IELLSLDDLLARADFISVHLPKTPETAGLIDKEALAKTKPGVIIVNAARG
MLBr_01692|M.leprae_Br4923 IELMSFDDLLARADFISVHLPKTPETAGLIDKEALAKTKPGVIIVNAARG
MAB_3304c|M.abscessus_ATCC_199 IELLTLDELLGRADFISVHLPKTPETAGLIGKEALAKTKPGVIIVNAARG
TH_2037|M.thermoresistible__bu IELVDLDELLGRADIISVHLPKTPETAGLIGADALAKVKPGVIIVNAARG
Mflv_5240|M.gilvum_PYR-GCK --FASPVELAASVDVLVVAAAGGDSSRGLVSSEVIEALGPSGYLINIARG
Mvan_1025|M.vanbaalenii_PYR-1 --AASPVELAASVDVLVVAAAGGDGTRGLVSSEVLDALGPHGYLINIARG
MSMEG_6126|M.smegmatis_MC2_155 --VGSAADLAAQVDVLIIAAAGGAGTRHLVDRAVLDALGPDGYLVNIARG
MAV_0123|M.avium_104 --AASAVELAELVDVLVVATTGDHQAHKLVDRAVLRALGPEGYLINIARG
:* .*.: : . : *:. .: * ::* ***
MMAR_1715|M.marinum_M GLIDESALAEAITSGHVRGAGLDVFATEPCTDSPLFELPQVVVTPHLGAS
MUL_1952|M.ulcerans_Agy99 GLIDESALAEAITSGYVRGAGLDVFATEPCTDSPLFELPQVVVTPHLGAS
Mb3020c|M.bovis_AF2122/97 GLVDEAALADAITGGHVRAAGLDVFATEPCTDSPLFELAQVVVTPHLGAS
Rv2996c|M.tuberculosis_H37Rv GLVDEAALADAITGGHVRAAGLDVFATEPCTDSPLFELAQVVVTPHLGAS
MLBr_01692|M.leprae_Br4923 GLVDEVALADAVRSGHVRAAGLDVFATEPCTDSPLFELSQVVVTPHLGAS
MAB_3304c|M.abscessus_ATCC_199 GLIDEAALADAIRSGHVRGAGLDVFSTEPCTDSPLFELDQVVVTPHLGAS
TH_2037|M.thermoresistible__bu GLIDEQALADAIISGRVRAAGIDVFAKEPCTDSPLFDLPQVVVTPHLGAS
Mflv_5240|M.gilvum_PYR-GCK SVVDQDALVVALVEKRLAGAGLDVFADEPHVPEELFALDNVVLLPHVGSG
Mvan_1025|M.vanbaalenii_PYR-1 SVVDQDALVSALVERRLAGAGLDVFADEPQVPEELFALDTVVLLPHVASG
MSMEG_6126|M.smegmatis_MC2_155 SVVDEDALVEALADGRLAGAGLDVFTDEPNVPEALLGMENVVLLPHVGSA
MAV_0123|M.avium_104 SVVDQEALVEMLAGGELAGAGLDVFADEPHVPAELVGLDNVVLLPHVGSA
.::*: **. : : .**:***: ** . *. : **: **:.:.
MMAR_1715|M.marinum_M TAEAQDRAGTDVAASVKLALAGEFVPDAVNIAGGAVNEEVAPWLDLACKL
MUL_1952|M.ulcerans_Agy99 TAEAQDRAGTDVAASVKLALAGEFVPDAVNIAGGAVNEEVAPWLDLACKL
Mb3020c|M.bovis_AF2122/97 TAEAQDRAGTDVAESVRLALAGEFVPDAVNVGGGVVNEEVAPWLDLVRKL
Rv2996c|M.tuberculosis_H37Rv TAEAQDRAGTDVAESVRLALAGEFVPDAVNVGGGVVNEEVAPWLDLVRKL
MLBr_01692|M.leprae_Br4923 TAEAQDRAGTDVAESVRLALAGEFVPDAVNVDGGVVNEEVAPWLDLVCKL
MAB_3304c|M.abscessus_ATCC_199 TSEAQDRAGTDVAASVQLALAGEFVPDAVNVGGGVVSEEVAPWLEVVRKL
TH_2037|M.thermoresistible__bu TVEAQDRAGTDVAASVKLALAGEFVPDAVNVGAGEVSEEVAPWVDLARKL
Mflv_5240|M.gilvum_PYR-GCK TVQTRAAMEELTVRNLHSFLTTGALVTPV---------------------
Mvan_1025|M.vanbaalenii_PYR-1 TVQTRAAMEALTLRNLDEFLATGELVTPVPLPAAAN--------------
MSMEG_6126|M.smegmatis_MC2_155 TVETRNAMEALTLANLDAYLKTGELVTPVPMPAAAPAPTPVQ--------
MAV_0123|M.avium_104 TARTRRAMASLALRNLDSYLATGQLVTPVLRPRRRIPDRN----------
* .:: . .: * : .*
MMAR_1715|M.marinum_M GVLAAALSDEPPVSLSVQARGELASEDVEVLKLSALRGLFSAVVEGPVTF
MUL_1952|M.ulcerans_Agy99 GVLAAALSDEPPVSLSVQAGGELASEDVEVLKLSALRGLFSAVVEGPVTF
Mb3020c|M.bovis_AF2122/97 GVLAGVLSDELPVSLSVQVRGELAAEEVEVLRLSALRGLFSAVIEDAVTF
Rv2996c|M.tuberculosis_H37Rv GVLAGVLSDELPVSLSVQVRGELAAEEVEVLRLSALRGLFSAVIEDAVTF
MLBr_01692|M.leprae_Br4923 GVLVAALSDELPASLSVHVRGELASEDVEILRLSALRGLFSTVIEDAVTF
MAB_3304c|M.abscessus_ATCC_199 GVLIGAVSEQLPTSLSVDVRGELASEDVAVLKLSALRGLFSSVIEDQVTF
TH_2037|M.thermoresistible__bu GLLVGTLSGEPTATLQVRVAGELASEDVEVLRLSALRGLFSTLTDQQVTY
Mflv_5240|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1025|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6126|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0123|M.avium_104 --------------------------------------------------
MMAR_1715|M.marinum_M VNAPALAAERGVSVEISKASESPNHRSVVDVRVVGADGSVVSVSGTLYGA
MUL_1952|M.ulcerans_Agy99 VNAPALAAERGVSVEISKASESPNHRSVVDVRVVGADGSVVSVSGTLYGA
Mb3020c|M.bovis_AF2122/97 VNAPALAAERGVTAEICKASESPNHRSVVDVRAVGADGSVVTVSGTLYGP
Rv2996c|M.tuberculosis_H37Rv VNAPALAAERGVTAEICKASESPNHRSVVDVRAVGADGSVVTVSGTLYGP
MLBr_01692|M.leprae_Br4923 VNAPALAAERGVSAEITTGSESPNHRSVVDVRAVASDGSVVNIAGTLSGP
MAB_3304c|M.abscessus_ATCC_199 VNAPSIAEERGVSAEITTATESPNHRSVVDVRAVYADGSVINVAGTLSGP
TH_2037|M.thermoresistible__bu VNAPTLAAERGMQAELEKVSESPNYRSVVDVRAVSTDGTVTNVAGTLSGP
Mflv_5240|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1025|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6126|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0123|M.avium_104 --------------------------------------------------
MMAR_1715|M.marinum_M QLTHKIVQINGRHFDLRAEGTNLIVNYVDQPGALGKIGTLLGAAGVNIHA
MUL_1952|M.ulcerans_Agy99 QLTHKIVQINGRHFDLRAEGTNLIVNYVDQPGALGKIGTLLGAAGVNIHA
Mb3020c|M.bovis_AF2122/97 QLSQKIVQINGRHFDLRAQGINLIIHYVDRPGALGKIGTLLGTAGVNIQA
Rv2996c|M.tuberculosis_H37Rv QLSQKIVQINGRHFDLRAQGINLIIHYVDRPGALGKIGTLLGTAGVNIQA
MLBr_01692|M.leprae_Br4923 QLVQKIVQVNGRNFDLRAQGMNLVIRYVDQPGALGKIGTLLGAAGVNIQA
MAB_3304c|M.abscessus_ATCC_199 QLVQKIVQINGRNFDLRAEGVNLVINYADVPGALGKIGTVLGGAEVNIQA
TH_2037|M.thermoresistible__bu QQVEKIVQINGRNFDLRAEGVNLVMHYVDRPGALGKIGTLLGAADVNILA
Mflv_5240|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1025|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6126|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0123|M.avium_104 --------------------------------------------------
MMAR_1715|M.marinum_M AQLSEDVEGPGATVLLRLDQDVPEDVRSAIAAAVGANRLEVVDLS
MUL_1952|M.ulcerans_Agy99 AQLSEDVEGPGATVLLRLDQDVPEDVRSAIAAAVGANRLEVVDLS
Mb3020c|M.bovis_AF2122/97 AQLSEDAEGPGATILLRLDQDVPDDVRTAIAAAVDAYKLEVVDLS
Rv2996c|M.tuberculosis_H37Rv AQLSEDAEGPGATILLRLDQDVPDDVRTAIAAAVDAYKLEVVDLS
MLBr_01692|M.leprae_Br4923 AQLSEDTEGPGATILLRLDQDVPGDVRSAIVAAVSANKLEVVNLS
MAB_3304c|M.abscessus_ATCC_199 AQLSQDASGAAATIILRIDRTAPDAVLDEIRAAVGATTLELVDLS
TH_2037|M.thermoresistible__bu AQLSQDISGDGATLMLRVNRDVPADVRDAIGAAVGASTLEVVDLS
Mflv_5240|M.gilvum_PYR-GCK ---------------------------------------------
Mvan_1025|M.vanbaalenii_PYR-1 ---------------------------------------------
MSMEG_6126|M.smegmatis_MC2_155 ---------------------------------------------
MAV_0123|M.avium_104 ---------------------------------------------