For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKVVVTRALPPATLSPLADVGEVWVSPHDRPLTDDELRHAVRGAHGIVSMLNDRIDERVLAAAGDQLRVV ANTAVGYDNLDVAAIARHGATATNTPGVLVDATADLTMALLLDVTRRVSEGDRLIRSGQPWSWDIGFMLG TGLQGKQLGIVGMGHIGRAVARRATAFGVRVVYHARRAQDDGIGRRVPLDELLATSDIVSLHCPLTIETR HLIDAEALGAMKPGSYLINTARGPIVDESALADALARGGIAGAALDVYEHEPEVHPGLRELPNVVLAPHL GSATVETRTLMAELAVKNVVQTLNDSGPVTPIAVPSG
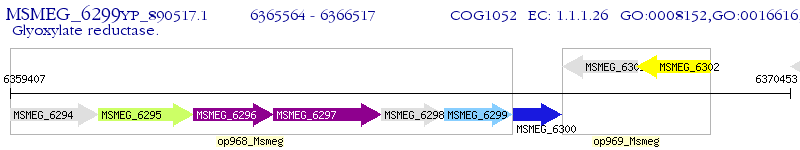
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6299 | - | - | 100% (317) | glyoxylate reductase |
| M. smegmatis MC2 155 | MSMEG_2529 | - | 2e-70 | 46.03% (315) | glyoxylate reductase |
| M. smegmatis MC2 155 | MSMEG_6126 | - | 9e-49 | 40.41% (292) | D-isomer specific 2-hydroxyacid dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 1e-47 | 38.94% (321) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4234 | - | 3e-48 | 39.13% (322) | D-3-phosphoglycerate dehydrogenase |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 2e-47 | 38.94% (321) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 1e-45 | 39.38% (292) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3304c | - | 5e-46 | 37.69% (321) | D-3-phosphoglycerate dehydrogenase |
| M. marinum M | MMAR_1715 | serA1 | 5e-47 | 39.73% (292) | D-3-phosphoglycerate dehydrogenase SerA1 |
| M. avium 104 | MAV_3847 | serA | 1e-46 | 39.38% (292) | D-3-phosphoglycerate dehydrogenase |
| M. thermoresistible (build 8) | TH_2037 | serA | 8e-46 | 37.07% (321) | PROBABLE D-3-PHOSPHOGLYCERATE DEHYDROGENASE SERA1 (PGDH) |
| M. ulcerans Agy99 | MUL_1952 | serA1 | 4e-47 | 39.73% (292) | D-3-phosphoglycerate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2128 | - | 6e-48 | 39.44% (322) | D-3-phosphoglycerate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3020c|M.bovis_AF2122/97 MSLPVVLIADKLAPSTVAALGDQVEVRWVDGPDR----DKLLAAVPEADA
Rv2996c|M.tuberculosis_H37Rv MSLPVVLIADKLAPSTVAALGDQVEVRWVDGPDR----DKLLAAVPEADA
MLBr_01692|M.leprae_Br4923 MDLPVVLIADKLAQSTVAALGDQVEVRWVDGPDR----TKLLAAVPEADA
MMAR_1715|M.marinum_M MNLPVVLIADKLAQSTVAALGDQVEVRWVDGPDR----EKLLAAVPEADA
MUL_1952|M.ulcerans_Agy99 MNLPVVLIADKLAQSTVAALGDQVEVRWVDGPDR----EKLLAAVPEADA
MAV_3847|M.avium_104 MNLPVVLIADKLAESTVAALGDQVEVRWVDGPDR----EKLLAAVPEADA
TH_2037|M.thermoresistible__bu VSLPVVLIADKLAESTVAALGDQVEVRWVDGPDR----EKLLAAVADADA
Mflv_4234|M.gilvum_PYR-GCK MSLPVVLIADKLAQSTVEALGDQVEVRWVDGPDR----EKLLAAVADADA
Mvan_2128|M.vanbaalenii_PYR-1 MSLPVVLIADKLAQSTVEALGDQVEVRWVDGPDR----EKLLAAVADADA
MAB_3304c|M.abscessus_ATCC_199 -----MLIADKLAPSTVEALGDGVEVRWVDGPDR----PALLAAVPEADA
MSMEG_6299|M.smegmatis_MC2_155 ---MKVVVTRALPPATLSPLADVGEV-WVSPHDRPLTDDELRHAVRGAHG
:::: *. :*: .*.* ** **. ** * ** *..
Mb3020c|M.bovis_AF2122/97 LLVRSATTVDAEVLAAA-PKLKIVARAGVGLDNVDVDAATARGVLVVNAP
Rv2996c|M.tuberculosis_H37Rv LLVRSATTVDAEVLAAA-PKLKIVARAGVGLDNVDVDAATARGVLVVNAP
MLBr_01692|M.leprae_Br4923 LLVRSATTVDAEVLAAA-PKLKIVARAGVGLDNVDVDAATARGVLVVNAP
MMAR_1715|M.marinum_M LLVRSATTVDAEVLAAA-TKLKIVARAGVGLDNVDVDAATTRGVLVVNAP
MUL_1952|M.ulcerans_Agy99 LLVRSATTVDAEVLAAA-TKLKIVARAGVGLDNVDVDAATTRGVLVVNAP
MAV_3847|M.avium_104 LLVRSATTVDAEVLAAA-PKLKIVARAGVGLDNVDVDAATERGVLVVNAP
TH_2037|M.thermoresistible__bu LLVRSATTVDAEVLAAA-PKLKIVARAGVGLDNVDVEAATARGVLVVNAP
Mflv_4234|M.gilvum_PYR-GCK LLVRSATTVDAEVLAAA-PKLKIVARAGVGLDNVDVDAATARGVLVVNAP
Mvan_2128|M.vanbaalenii_PYR-1 LLVRSATTVDAEVLAAA-PKLKIVARAGVGLDNVDVDAATARGVLVVNAP
MAB_3304c|M.abscessus_ATCC_199 LLVRSATTVDAEVLAAG-TKLKIVARAGVGLDNVDVKAATARGVLVVNAP
MSMEG_6299|M.smegmatis_MC2_155 IVSMLNDRIDERVLAAAGDQLRVVANTAVGYDNLDVAAIARHGATATNTP
:: :* .****. :*::**.:.** **:** * : :*. ..*:*
Mb3020c|M.bovis_AF2122/97 TSNIHSAAEHALALLLAASRQIPAADASLR---EHTWKRSSFSGTEIFGK
Rv2996c|M.tuberculosis_H37Rv TSNIHSAAEHALALLLAASRQIPAADASLR---EHTWKRSSFSGTEIFGK
MLBr_01692|M.leprae_Br4923 TSNIHSAAEHALALLLAASRQIAEADASLR---AHIWKRSSFSGTEIFGK
MMAR_1715|M.marinum_M TSNIHSAAEHAMALLLAAARQIPAADASLR---EHTWKRSSFSGAEIFGK
MUL_1952|M.ulcerans_Agy99 TSNIHSAAEHAMALLLAAARQIPAADASLR---EHTWKRSSFSGAEIFGK
MAV_3847|M.avium_104 TSNIHSAAEHALALLLAAAREIPAADASLR---EHTWKRSSFSGTEIFGK
TH_2037|M.thermoresistible__bu TSNIHSAAEHAIALLLAAARQIPAADATLR---QRTWKRSEFSGVEIYDK
Mflv_4234|M.gilvum_PYR-GCK TSNIHSAAEHALALLLSTARQIPAADATLR---EHTWKRSAFSGTEIFGK
Mvan_2128|M.vanbaalenii_PYR-1 TSNIHSAAEHALALLLAAARQIPAADATLR---EHSWKRSSFSGTEIFGK
MAB_3304c|M.abscessus_ATCC_199 TSNIHSAAEHAVTLLLATARQIPAADASLK---AHTWKRSSFNGTEIFGK
MSMEG_6299|M.smegmatis_MC2_155 GVLVDATADLTMALLLDVTRRVSEGDRLIRSGQPWSWDIGFMLGTGLQGK
:.::*: :::*** .:*.:. .* :: *. . : *. : .*
Mb3020c|M.bovis_AF2122/97 TVGVVGLGRIGQLVAQRIAAFGAYVVAYDPYVSPARAAQLGIELLSLDDL
Rv2996c|M.tuberculosis_H37Rv TVGVVGLGRIGQLVAQRIAAFGAYVVAYDPYVSPARAAQLGIELLSLDDL
MLBr_01692|M.leprae_Br4923 TVGVVGLGRIGQLVAARIAAFGAHVIAYDPYVAPARAAQLGIELMSFDDL
MMAR_1715|M.marinum_M TVGVVGLGRIGQLVAQRVEAFGAHVVAYDPYVSPARAAQLGIELLPLDDL
MUL_1952|M.ulcerans_Agy99 TVGVVGLGRIGQLVAQRVEAFGAHVVAYDPYVSPARAAQLGIELLPLDDL
MAV_3847|M.avium_104 TVGVVGLGRIGQLVAQRLSAFGTHLIAYDPYVSPARAAQLGIELLSLDEL
TH_2037|M.thermoresistible__bu TVGVVGLGRIGQLVAQRLAAFGTHLIAYDPYVSPARAAQLGIELVDLDEL
Mflv_4234|M.gilvum_PYR-GCK TVGVVGLGRIGQLVAQRLAAFGAHITAYDPYVSHARAAQLGIELLTLDEL
Mvan_2128|M.vanbaalenii_PYR-1 TVGVVGLGRIGQLVAQRLAAFGAHITAYDPYVSHARAAQLGIELLTLDEL
MAB_3304c|M.abscessus_ATCC_199 TVGVVGLGRIGQLFAQRLAAFGTHIVAYDPYVSAARAAQLGIELLTLDEL
MSMEG_6299|M.smegmatis_MC2_155 QLGIVGMGHIGRAVARRATAFGVRVVYHARRAQDDGIGRR----VPLDEL
:*:**:*:**: .* * ***. : : . .: : :*:*
Mb3020c|M.bovis_AF2122/97 LARADFISVHLPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALA
Rv2996c|M.tuberculosis_H37Rv LARADFISVHLPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALA
MLBr_01692|M.leprae_Br4923 LARADFISVHLPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEVALA
MMAR_1715|M.marinum_M LARADFISVHLPKTPETAGLLGKEALAKTKPGVIIVNAARGGLIDESALA
MUL_1952|M.ulcerans_Agy99 LARADFISVHLPKTPETAGLLGKEALAKTKPGVIIVNAARGGLIDESALA
MAV_3847|M.avium_104 LARADFISVHLPKTPETAGLIDKDALAKTKPGVIIVNAARGGLVDEAALA
TH_2037|M.thermoresistible__bu LGRADIISVHLPKTPETAGLIGADALAKVKPGVIIVNAARGGLIDEQALA
Mflv_4234|M.gilvum_PYR-GCK LSRADFISVHLPKTKETAGLIGTEALAKTKPGVIIVNAARGGLIDEQALA
Mvan_2128|M.vanbaalenii_PYR-1 LGRADFISVHLPKTKETAGLIGKEALAKTKPGVIIVNAARGGLIDEAALA
MAB_3304c|M.abscessus_ATCC_199 LGRADFISVHLPKTPETAGLIGKEALAKTKPGVIIVNAARGGLIDEAALA
MSMEG_6299|M.smegmatis_MC2_155 LATSDIVSLHCPLTIETRHLIDAEALGAMKPGSYLINTARGPIVDESALA
*. :*::*:* * * ** *:. :**. *** ::*:*** ::** ***
Mb3020c|M.bovis_AF2122/97 DAITGGHVRAAGLDVFATEPCTDSPLFELAQVVVTPHLGASTAEAQDRAG
Rv2996c|M.tuberculosis_H37Rv DAITGGHVRAAGLDVFATEPCTDSPLFELAQVVVTPHLGASTAEAQDRAG
MLBr_01692|M.leprae_Br4923 DAVRSGHVRAAGLDVFATEPCTDSPLFELSQVVVTPHLGASTAEAQDRAG
MMAR_1715|M.marinum_M EAITSGHVRGAGLDVFATEPCTDSPLFELPQVVVTPHLGASTAEAQDRAG
MUL_1952|M.ulcerans_Agy99 EAITSGYVRGAGLDVFATEPCTDSPLFELPQVVVTPHLGASTAEAQDRAG
MAV_3847|M.avium_104 DAVRSGHVRAAGLDVFSKEPCTDSPLFELPQVVVTPHLGASTTEAQDRAG
TH_2037|M.thermoresistible__bu DAIISGRVRAAGIDVFAKEPCTDSPLFDLPQVVVTPHLGASTVEAQDRAG
Mflv_4234|M.gilvum_PYR-GCK DAITSGHVRGAGLDVFSTEPCTDSPLFELPQVVVTPHLGASTVEAQDRAG
Mvan_2128|M.vanbaalenii_PYR-1 DAINSGHVRGAGLDVFSTEPCTDSPLFELPQVVVTPHLGASTVEAQDRAG
MAB_3304c|M.abscessus_ATCC_199 DAIRSGHVRGAGLDVFSTEPCTDSPLFELDQVVVTPHLGASTSEAQDRAG
MSMEG_6299|M.smegmatis_MC2_155 DALARGGIAGAALDVYEHEPEVHPGLRELPNVVLAPHLGSATVETRTLMA
:*: * : .*.:**: ** ... * :* :**::****::* *:: .
Mb3020c|M.bovis_AF2122/97 TDVAESVRLALAGEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSD
Rv2996c|M.tuberculosis_H37Rv TDVAESVRLALAGEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSD
MLBr_01692|M.leprae_Br4923 TDVAESVRLALAGEFVPDAVNVDGGVVNEEVAPWLDLVCKLGVLVAALSD
MMAR_1715|M.marinum_M TDVAASVKLALAGEFVPDAVNIAGGAVNEEVAPWLDLACKLGVLAAALSD
MUL_1952|M.ulcerans_Agy99 TDVAASVKLALAGEFVPDAVNIAGGAVNEEVAPWLDLACKLGVLAAALSD
MAV_3847|M.avium_104 TDVAASVKLALAGEFVPDAVNVAGGTVNEEVAPWLDLVRKLGLLAATLSD
TH_2037|M.thermoresistible__bu TDVAASVKLALAGEFVPDAVNVGAGEVSEEVAPWVDLARKLGLLVGTLSG
Mflv_4234|M.gilvum_PYR-GCK TDVAASVKLALAGEFVPDAVNVGGGAVGEEVAPWLDLVRKLGLLVGALSA
Mvan_2128|M.vanbaalenii_PYR-1 TDVAASVKLALAGEFVPDAVNVGGGAVGEEVAPWLDLVRKLGLLVGVLSS
MAB_3304c|M.abscessus_ATCC_199 TDVAASVQLALAGEFVPDAVNVGGGVVSEEVAPWLEVVRKLGVLIGAVSE
MSMEG_6299|M.smegmatis_MC2_155 ELAVKNVVQTLNDSGPVTPIAVPSG-------------------------
.. .* :* .. .: : .*
Mb3020c|M.bovis_AF2122/97 ELPVSLSVQVRGELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAE
Rv2996c|M.tuberculosis_H37Rv ELPVSLSVQVRGELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAE
MLBr_01692|M.leprae_Br4923 ELPASLSVHVRGELASEDVEILRLSALRGLFSTVIEDAVTFVNAPALAAE
MMAR_1715|M.marinum_M EPPVSLSVQARGELASEDVEVLKLSALRGLFSAVVEGPVTFVNAPALAAE
MUL_1952|M.ulcerans_Agy99 EPPVSLSVQAGGELASEDVEVLKLSALRGLFSAVVEGPVTFVNAPALAAE
MAV_3847|M.avium_104 GPPVSLSVQVRGELASEDVEVLKLSALRGLFSAVVEGPVTFVNAPALAEE
TH_2037|M.thermoresistible__bu EPTATLQVRVAGELASEDVEVLRLSALRGLFSTLTDQQVTYVNAPTLAAE
Mflv_4234|M.gilvum_PYR-GCK ELPTNLCVQVRGELASEEVEVLRLSALRGLFSAVIEDQVTFVNAPALATE
Mvan_2128|M.vanbaalenii_PYR-1 EPPVSLQVQVQGELASEEVEVLKLSALRGLFSAVIEHPVTFVNAPALASE
MAB_3304c|M.abscessus_ATCC_199 QLPTSLSVDVRGELASEDVAVLKLSALRGLFSSVIEDQVTFVNAPSIAEE
MSMEG_6299|M.smegmatis_MC2_155 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 RGVTAEICKASESPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQI
Rv2996c|M.tuberculosis_H37Rv RGVTAEICKASESPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQI
MLBr_01692|M.leprae_Br4923 RGVSAEITTGSESPNHRSVVDVRAVASDGSVVNIAGTLSGPQLVQKIVQV
MMAR_1715|M.marinum_M RGVSVEISKASESPNHRSVVDVRVVGADGSVVSVSGTLYGAQLTHKIVQI
MUL_1952|M.ulcerans_Agy99 RGVSVEISKASESPNHRSVVDVRVVGADGSVVSVSGTLYGAQLTHKIVQI
MAV_3847|M.avium_104 RGVTAEISTASESPNHRSVVDVRAVSHDGTVVNVAGTLSGPQLVEKIVQI
TH_2037|M.thermoresistible__bu RGMQAELEKVSESPNYRSVVDVRAVSTDGTVTNVAGTLSGPQQVEKIVQI
Mflv_4234|M.gilvum_PYR-GCK RGVEASIDTESESPNHRSVVDVRAVAADGSTVNVSGTLTGPQLVEKIVQI
Mvan_2128|M.vanbaalenii_PYR-1 RGVEASITTATESANHRSVVDVRAVAADGSTVNVAGTLTGPQLVEKIVQI
MAB_3304c|M.abscessus_ATCC_199 RGVSAEITTATESPNHRSVVDVRAVYADGSVINVAGTLSGPQLVQKIVQI
MSMEG_6299|M.smegmatis_MC2_155 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 NGRHFDLRAQGINLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEG
Rv2996c|M.tuberculosis_H37Rv NGRHFDLRAQGINLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEG
MLBr_01692|M.leprae_Br4923 NGRNFDLRAQGMNLVIRYVDQPGALGKIGTLLGAAGVNIQAAQLSEDTEG
MMAR_1715|M.marinum_M NGRHFDLRAEGTNLIVNYVDQPGALGKIGTLLGAAGVNIHAAQLSEDVEG
MUL_1952|M.ulcerans_Agy99 NGRHFDLRAEGTNLIVNYVDQPGALGKIGTLLGAAGVNIHAAQLSEDVEG
MAV_3847|M.avium_104 NGRNFDLRARGINLVINYVDQPGALGKIGTLLGSAGVNIQAAQLSEDAEG
TH_2037|M.thermoresistible__bu NGRNFDLRAEGVNLVMHYVDRPGALGKIGTLLGAADVNILAAQLSQDISG
Mflv_4234|M.gilvum_PYR-GCK NGRNLDLRAEGVNVIINYHDQPGALGKIGTLLGGANVNILAAQLSQDADG
Mvan_2128|M.vanbaalenii_PYR-1 NGRNLELRAEGVNLIINYDDQPGALGKIGTLLGGAAVNILAAQLSQDADG
MAB_3304c|M.abscessus_ATCC_199 NGRNFDLRAEGVNLVINYADVPGALGKIGTVLGGAEVNIQAAQLSQDASG
MSMEG_6299|M.smegmatis_MC2_155 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 PGATILLRLDQDVPDDVRTAIAAAVDAYKLEVVDLS
Rv2996c|M.tuberculosis_H37Rv PGATILLRLDQDVPDDVRTAIAAAVDAYKLEVVDLS
MLBr_01692|M.leprae_Br4923 PGATILLRLDQDVPGDVRSAIVAAVSANKLEVVNLS
MMAR_1715|M.marinum_M PGATVLLRLDQDVPEDVRSAIAAAVGANRLEVVDLS
MUL_1952|M.ulcerans_Agy99 PGATVLLRLDQDVPEDVRSAIAAAVGANRLEVVDLS
MAV_3847|M.avium_104 PGATILLRLDQDVPADVRSAIGAAVGANKLEVVDLS
TH_2037|M.thermoresistible__bu DGATLMLRVNRDVPADVRDAIGAAVGASTLEVVDLS
Mflv_4234|M.gilvum_PYR-GCK EGATIMLRVDREVPADVLAGIGRDVNALTLEVVDLT
Mvan_2128|M.vanbaalenii_PYR-1 IGATVMLRLDREVPGEVLAAIGRDVNAVTLEVVDLT
MAB_3304c|M.abscessus_ATCC_199 AAATIILRIDRTAPDAVLDEIRAAVGATTLELVDLS
MSMEG_6299|M.smegmatis_MC2_155 ------------------------------------