For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNLPVVLIADKLAQSTVAALGDQVEVRWVDGPDREKLLAAVPEADALLVRSATTVDAEVLAAATKLKIVA RAGVGLDNVDVDAATTRGVLVVNAPTSNIHSAAEHAMALLLAAARQIPAADASLREHTWKRSSFSGAEIF GKTVGVVGLGRIGQLVAQRVEAFGAHVVAYDPYVSPARAAQLGIELLPLDDLLARADFISVHLPKTPETA GLLGKEALAKTKPGVIIVNAARGGLIDESALAEAITSGYVRGAGLDVFATEPCTDSPLFELPQVVVTPHL GASTAEAQDRAGTDVAASVKLALAGEFVPDAVNIAGGAVNEEVAPWLDLACKLGVLAAALSDEPPVSLSV QAGGELASEDVEVLKLSALRGLFSAVVEGPVTFVNAPALAAERGVSVEISKASESPNHRSVVDVRVVGAD GSVVSVSGTLYGAQLTHKIVQINGRHFDLRAEGTNLIVNYVDQPGALGKIGTLLGAAGVNIHAAQLSEDV EGPGATVLLRLDQDVPEDVRSAIAAAVGANRLEVVDLS
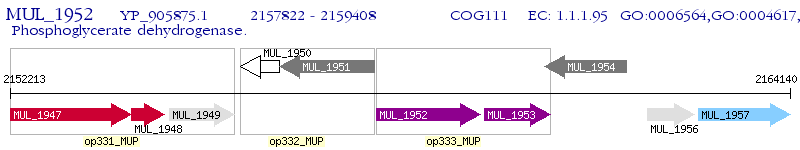
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1952 | serA1 | - | 100% (528) | D-3-phosphoglycerate dehydrogenase |
| M. ulcerans Agy99 | MUL_1195 | serA3 | 4e-42 | 35.86% (304) | D-3-phosphoglycerate dehydrogenase SerA3 |
| M. ulcerans Agy99 | MUL_0823 | serA2 | 2e-40 | 35.58% (312) | D-3-phosphoglycerate dehydrogenase SerA2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 0.0 | 88.45% (528) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4234 | - | 0.0 | 82.58% (528) | D-3-phosphoglycerate dehydrogenase |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 0.0 | 88.45% (528) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 0.0 | 85.61% (528) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3304c | - | 0.0 | 80.50% (523) | D-3-phosphoglycerate dehydrogenase |
| M. marinum M | MMAR_1715 | serA1 | 0.0 | 99.62% (528) | D-3-phosphoglycerate dehydrogenase SerA1 |
| M. avium 104 | MAV_3847 | serA | 0.0 | 88.45% (528) | D-3-phosphoglycerate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2378 | serA | 0.0 | 85.23% (528) | D-3-phosphoglycerate dehydrogenase |
| M. thermoresistible (build 8) | TH_2037 | serA | 0.0 | 79.73% (528) | PROBABLE D-3-PHOSPHOGLYCERATE DEHYDROGENASE SERA1 (PGDH) |
| M. vanbaalenii PYR-1 | Mvan_2128 | - | 0.0 | 83.71% (528) | D-3-phosphoglycerate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_1952|M.ulcerans_Agy99 MNLPVVLIADKLAQSTVAALGDQVEVRWVDGPDREKLLAAVPEADALLVR
MMAR_1715|M.marinum_M MNLPVVLIADKLAQSTVAALGDQVEVRWVDGPDREKLLAAVPEADALLVR
Mb3020c|M.bovis_AF2122/97 MSLPVVLIADKLAPSTVAALGDQVEVRWVDGPDRDKLLAAVPEADALLVR
Rv2996c|M.tuberculosis_H37Rv MSLPVVLIADKLAPSTVAALGDQVEVRWVDGPDRDKLLAAVPEADALLVR
MLBr_01692|M.leprae_Br4923 MDLPVVLIADKLAQSTVAALGDQVEVRWVDGPDRTKLLAAVPEADALLVR
MAV_3847|M.avium_104 MNLPVVLIADKLAESTVAALGDQVEVRWVDGPDREKLLAAVPEADALLVR
Mflv_4234|M.gilvum_PYR-GCK MSLPVVLIADKLAQSTVEALGDQVEVRWVDGPDREKLLAAVADADALLVR
Mvan_2128|M.vanbaalenii_PYR-1 MSLPVVLIADKLAQSTVEALGDQVEVRWVDGPDREKLLAAVADADALLVR
MAB_3304c|M.abscessus_ATCC_199 -----MLIADKLAPSTVEALGDGVEVRWVDGPDRPALLAAVPEADALLVR
MSMEG_2378|M.smegmatis_MC2_155 MSLPVVLIADKLAESTVAALGDEVEVRWVDGPDRAKLLAAVPEADALLVR
TH_2037|M.thermoresistible__bu VSLPVVLIADKLAESTVAALGDQVEVRWVDGPDREKLLAAVADADALLVR
:******* *** **** *********** *****.:*******
MUL_1952|M.ulcerans_Agy99 SATTVDAEVLAAATKLKIVARAGVGLDNVDVDAATTRGVLVVNAPTSNIH
MMAR_1715|M.marinum_M SATTVDAEVLAAATKLKIVARAGVGLDNVDVDAATTRGVLVVNAPTSNIH
Mb3020c|M.bovis_AF2122/97 SATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAATARGVLVVNAPTSNIH
Rv2996c|M.tuberculosis_H37Rv SATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAATARGVLVVNAPTSNIH
MLBr_01692|M.leprae_Br4923 SATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAATARGVLVVNAPTSNIH
MAV_3847|M.avium_104 SATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAATERGVLVVNAPTSNIH
Mflv_4234|M.gilvum_PYR-GCK SATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAATARGVLVVNAPTSNIH
Mvan_2128|M.vanbaalenii_PYR-1 SATTVDAEVLAAAPKLKIVARAGVGLDNVDVDAATARGVLVVNAPTSNIH
MAB_3304c|M.abscessus_ATCC_199 SATTVDAEVLAAGTKLKIVARAGVGLDNVDVKAATARGVLVVNAPTSNIH
MSMEG_2378|M.smegmatis_MC2_155 SATTVDAEVIAAAPKLKIVARAGVGLDNVDVDAATARGVLVVNAPTSNIH
TH_2037|M.thermoresistible__bu SATTVDAEVLAAAPKLKIVARAGVGLDNVDVEAATARGVLVVNAPTSNIH
*********:**..*****************.*** **************
MUL_1952|M.ulcerans_Agy99 SAAEHAMALLLAAARQIPAADASLREHTWKRSSFSGAEIFGKTVGVVGLG
MMAR_1715|M.marinum_M SAAEHAMALLLAAARQIPAADASLREHTWKRSSFSGAEIFGKTVGVVGLG
Mb3020c|M.bovis_AF2122/97 SAAEHALALLLAASRQIPAADASLREHTWKRSSFSGTEIFGKTVGVVGLG
Rv2996c|M.tuberculosis_H37Rv SAAEHALALLLAASRQIPAADASLREHTWKRSSFSGTEIFGKTVGVVGLG
MLBr_01692|M.leprae_Br4923 SAAEHALALLLAASRQIAEADASLRAHIWKRSSFSGTEIFGKTVGVVGLG
MAV_3847|M.avium_104 SAAEHALALLLAAAREIPAADASLREHTWKRSSFSGTEIFGKTVGVVGLG
Mflv_4234|M.gilvum_PYR-GCK SAAEHALALLLSTARQIPAADATLREHTWKRSAFSGTEIFGKTVGVVGLG
Mvan_2128|M.vanbaalenii_PYR-1 SAAEHALALLLAAARQIPAADATLREHSWKRSSFSGTEIFGKTVGVVGLG
MAB_3304c|M.abscessus_ATCC_199 SAAEHAVTLLLATARQIPAADASLKAHTWKRSSFNGTEIFGKTVGVVGLG
MSMEG_2378|M.smegmatis_MC2_155 SAAEHAIALLLATARQIPAADATLRERSWKRSSFSGTEIFGKTVGVVGLG
TH_2037|M.thermoresistible__bu SAAEHAIALLLAAARQIPAADATLRQRTWKRSEFSGVEIYDKTVGVVGLG
******::***:::*:*. ***:*: : **** *.*.**:.*********
MUL_1952|M.ulcerans_Agy99 RIGQLVAQRVEAFGAHVVAYDPYVSPARAAQLGIELLPLDDLLARADFIS
MMAR_1715|M.marinum_M RIGQLVAQRVEAFGAHVVAYDPYVSPARAAQLGIELLPLDDLLARADFIS
Mb3020c|M.bovis_AF2122/97 RIGQLVAQRIAAFGAYVVAYDPYVSPARAAQLGIELLSLDDLLARADFIS
Rv2996c|M.tuberculosis_H37Rv RIGQLVAQRIAAFGAYVVAYDPYVSPARAAQLGIELLSLDDLLARADFIS
MLBr_01692|M.leprae_Br4923 RIGQLVAARIAAFGAHVIAYDPYVAPARAAQLGIELMSFDDLLARADFIS
MAV_3847|M.avium_104 RIGQLVAQRLSAFGTHLIAYDPYVSPARAAQLGIELLSLDELLARADFIS
Mflv_4234|M.gilvum_PYR-GCK RIGQLVAQRLAAFGAHITAYDPYVSHARAAQLGIELLTLDELLSRADFIS
Mvan_2128|M.vanbaalenii_PYR-1 RIGQLVAQRLAAFGAHITAYDPYVSHARAAQLGIELLTLDELLGRADFIS
MAB_3304c|M.abscessus_ATCC_199 RIGQLFAQRLAAFGTHIVAYDPYVSAARAAQLGIELLTLDELLGRADFIS
MSMEG_2378|M.smegmatis_MC2_155 RIGQLVAQRLAAFGAHIVAYDPYVSQARAAQLGIELLPLDDLLARADFIS
TH_2037|M.thermoresistible__bu RIGQLVAQRLAAFGTHLIAYDPYVSPARAAQLGIELVDLDELLGRADIIS
*****.* *: ***::: ******: **********: :*:**.***:**
MUL_1952|M.ulcerans_Agy99 VHLPKTPETAGLLGKEALAKTKPGVIIVNAARGGLIDESALAEAITSGYV
MMAR_1715|M.marinum_M VHLPKTPETAGLLGKEALAKTKPGVIIVNAARGGLIDESALAEAITSGHV
Mb3020c|M.bovis_AF2122/97 VHLPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHV
Rv2996c|M.tuberculosis_H37Rv VHLPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHV
MLBr_01692|M.leprae_Br4923 VHLPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEVALADAVRSGHV
MAV_3847|M.avium_104 VHLPKTPETAGLIDKDALAKTKPGVIIVNAARGGLVDEAALADAVRSGHV
Mflv_4234|M.gilvum_PYR-GCK VHLPKTKETAGLIGTEALAKTKPGVIIVNAARGGLIDEQALADAITSGHV
Mvan_2128|M.vanbaalenii_PYR-1 VHLPKTKETAGLIGKEALAKTKPGVIIVNAARGGLIDEAALADAINSGHV
MAB_3304c|M.abscessus_ATCC_199 VHLPKTPETAGLIGKEALAKTKPGVIIVNAARGGLIDEAALADAIRSGHV
MSMEG_2378|M.smegmatis_MC2_155 VHLPKTKETAGLLGKEALAKTKKGVIIVNAARGGLIDEQALADAITSGHV
TH_2037|M.thermoresistible__bu VHLPKTPETAGLIGADALAKVKPGVIIVNAARGGLIDEQALADAIISGRV
****** *****:. :****.* ************:** ***:*: .* *
MUL_1952|M.ulcerans_Agy99 RGAGLDVFATEPCTDSPLFELPQVVVTPHLGASTAEAQDRAGTDVAASVK
MMAR_1715|M.marinum_M RGAGLDVFATEPCTDSPLFELPQVVVTPHLGASTAEAQDRAGTDVAASVK
Mb3020c|M.bovis_AF2122/97 RAAGLDVFATEPCTDSPLFELAQVVVTPHLGASTAEAQDRAGTDVAESVR
Rv2996c|M.tuberculosis_H37Rv RAAGLDVFATEPCTDSPLFELAQVVVTPHLGASTAEAQDRAGTDVAESVR
MLBr_01692|M.leprae_Br4923 RAAGLDVFATEPCTDSPLFELSQVVVTPHLGASTAEAQDRAGTDVAESVR
MAV_3847|M.avium_104 RAAGLDVFSKEPCTDSPLFELPQVVVTPHLGASTTEAQDRAGTDVAASVK
Mflv_4234|M.gilvum_PYR-GCK RGAGLDVFSTEPCTDSPLFELPQVVVTPHLGASTVEAQDRAGTDVAASVK
Mvan_2128|M.vanbaalenii_PYR-1 RGAGLDVFSTEPCTDSPLFELPQVVVTPHLGASTVEAQDRAGTDVAASVK
MAB_3304c|M.abscessus_ATCC_199 RGAGLDVFSTEPCTDSPLFELDQVVVTPHLGASTSEAQDRAGTDVAASVQ
MSMEG_2378|M.smegmatis_MC2_155 RGAGLDVYATEPCTDSPLFDLPQVVVTPHLGASTAEAQDRAGTDVAASVK
TH_2037|M.thermoresistible__bu RAAGIDVFAKEPCTDSPLFDLPQVVVTPHLGASTVEAQDRAGTDVAASVK
*.**:**::.*********:* ************ *********** **:
MUL_1952|M.ulcerans_Agy99 LALAGEFVPDAVNIAGGAVNEEVAPWLDLACKLGVLAAALSDEPPVSLSV
MMAR_1715|M.marinum_M LALAGEFVPDAVNIAGGAVNEEVAPWLDLACKLGVLAAALSDEPPVSLSV
Mb3020c|M.bovis_AF2122/97 LALAGEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELPVSLSV
Rv2996c|M.tuberculosis_H37Rv LALAGEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELPVSLSV
MLBr_01692|M.leprae_Br4923 LALAGEFVPDAVNVDGGVVNEEVAPWLDLVCKLGVLVAALSDELPASLSV
MAV_3847|M.avium_104 LALAGEFVPDAVNVAGGTVNEEVAPWLDLVRKLGLLAATLSDGPPVSLSV
Mflv_4234|M.gilvum_PYR-GCK LALAGEFVPDAVNVGGGAVGEEVAPWLDLVRKLGLLVGALSAELPTNLCV
Mvan_2128|M.vanbaalenii_PYR-1 LALAGEFVPDAVNVGGGAVGEEVAPWLDLVRKLGLLVGVLSSEPPVSLQV
MAB_3304c|M.abscessus_ATCC_199 LALAGEFVPDAVNVGGGVVSEEVAPWLEVVRKLGVLIGAVSEQLPTSLSV
MSMEG_2378|M.smegmatis_MC2_155 LALAGEFVPDAVNVGGGVVGEEVAPWLELVRKLGLLVGALADAPPVSLQV
TH_2037|M.thermoresistible__bu LALAGEFVPDAVNVGAGEVSEEVAPWVDLARKLGLLVGTLSGEPTATLQV
*************: .* *.******:::. ***:* ..:: ...* *
MUL_1952|M.ulcerans_Agy99 QAGGELASEDVEVLKLSALRGLFSAVVEGPVTFVNAPALAAERGVSVEIS
MMAR_1715|M.marinum_M QARGELASEDVEVLKLSALRGLFSAVVEGPVTFVNAPALAAERGVSVEIS
Mb3020c|M.bovis_AF2122/97 QVRGELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAERGVTAEIC
Rv2996c|M.tuberculosis_H37Rv QVRGELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAERGVTAEIC
MLBr_01692|M.leprae_Br4923 HVRGELASEDVEILRLSALRGLFSTVIEDAVTFVNAPALAAERGVSAEIT
MAV_3847|M.avium_104 QVRGELASEDVEVLKLSALRGLFSAVVEGPVTFVNAPALAEERGVTAEIS
Mflv_4234|M.gilvum_PYR-GCK QVRGELASEEVEVLRLSALRGLFSAVIEDQVTFVNAPALATERGVEASID
Mvan_2128|M.vanbaalenii_PYR-1 QVQGELASEEVEVLKLSALRGLFSAVIEHPVTFVNAPALASERGVEASIT
MAB_3304c|M.abscessus_ATCC_199 DVRGELASEDVAVLKLSALRGLFSSVIEDQVTFVNAPSIAEERGVSAEIT
MSMEG_2378|M.smegmatis_MC2_155 QVRGELASEDVEILKLSALRGLFSAVVDEQVTFVNAPALAADRGVTAEIS
TH_2037|M.thermoresistible__bu RVAGELASEDVEVLRLSALRGLFSTLTDQQVTYVNAPTLAAERGMQAELE
. ****:*:* :*:*********:: : **:****::* :**: ..:
MUL_1952|M.ulcerans_Agy99 KASESPNHRSVVDVRVVGADGSVVSVSGTLYGAQLTHKIVQINGRHFDLR
MMAR_1715|M.marinum_M KASESPNHRSVVDVRVVGADGSVVSVSGTLYGAQLTHKIVQINGRHFDLR
Mb3020c|M.bovis_AF2122/97 KASESPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGRHFDLR
Rv2996c|M.tuberculosis_H37Rv KASESPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGRHFDLR
MLBr_01692|M.leprae_Br4923 TGSESPNHRSVVDVRAVASDGSVVNIAGTLSGPQLVQKIVQVNGRNFDLR
MAV_3847|M.avium_104 TASESPNHRSVVDVRAVSHDGTVVNVAGTLSGPQLVEKIVQINGRNFDLR
Mflv_4234|M.gilvum_PYR-GCK TESESPNHRSVVDVRAVAADGSTVNVSGTLTGPQLVEKIVQINGRNLDLR
Mvan_2128|M.vanbaalenii_PYR-1 TATESANHRSVVDVRAVAADGSTVNVAGTLTGPQLVEKIVQINGRNLELR
MAB_3304c|M.abscessus_ATCC_199 TATESPNHRSVVDVRAVYADGSVINVAGTLSGPQLVQKIVQINGRNFDLR
MSMEG_2378|M.smegmatis_MC2_155 TATESPNHRSVVDVRAVHADGSSINVAGTLSGPQLVEKIVQINGRNFDLR
TH_2037|M.thermoresistible__bu KVSESPNYRSVVDVRAVSTDGTVTNVAGTLSGPQQVEKIVQINGRNFDLR
. :**.*:*******.* **: .::*** *.* .****:***:::**
MUL_1952|M.ulcerans_Agy99 AEGTNLIVNYVDQPGALGKIGTLLGAAGVNIHAAQLSEDVEGPGATVLLR
MMAR_1715|M.marinum_M AEGTNLIVNYVDQPGALGKIGTLLGAAGVNIHAAQLSEDVEGPGATVLLR
Mb3020c|M.bovis_AF2122/97 AQGINLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGATILLR
Rv2996c|M.tuberculosis_H37Rv AQGINLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGATILLR
MLBr_01692|M.leprae_Br4923 AQGMNLVIRYVDQPGALGKIGTLLGAAGVNIQAAQLSEDTEGPGATILLR
MAV_3847|M.avium_104 ARGINLVINYVDQPGALGKIGTLLGSAGVNIQAAQLSEDAEGPGATILLR
Mflv_4234|M.gilvum_PYR-GCK AEGVNVIINYHDQPGALGKIGTLLGGANVNILAAQLSQDADGEGATIMLR
Mvan_2128|M.vanbaalenii_PYR-1 AEGVNLIINYDDQPGALGKIGTLLGGAAVNILAAQLSQDADGIGATVMLR
MAB_3304c|M.abscessus_ATCC_199 AEGVNLVINYADVPGALGKIGTVLGGAEVNIQAAQLSQDASGAAATIILR
MSMEG_2378|M.smegmatis_MC2_155 AEGHNLIVNYSDKPGALGKIGTLLGGANVNILAAQLSQDASGDHATVMLR
TH_2037|M.thermoresistible__bu AEGVNLVMHYVDRPGALGKIGTLLGAADVNILAAQLSQDISGDGATLMLR
*.* *:::.* * *********:** * *** *****:* .* **::**
MUL_1952|M.ulcerans_Agy99 LDQDVPEDVRSAIAAAVGANRLEVVDLS
MMAR_1715|M.marinum_M LDQDVPEDVRSAIAAAVGANRLEVVDLS
Mb3020c|M.bovis_AF2122/97 LDQDVPDDVRTAIAAAVDAYKLEVVDLS
Rv2996c|M.tuberculosis_H37Rv LDQDVPDDVRTAIAAAVDAYKLEVVDLS
MLBr_01692|M.leprae_Br4923 LDQDVPGDVRSAIVAAVSANKLEVVNLS
MAV_3847|M.avium_104 LDQDVPADVRSAIGAAVGANKLEVVDLS
Mflv_4234|M.gilvum_PYR-GCK VDREVPADVLAGIGRDVNALTLEVVDLT
Mvan_2128|M.vanbaalenii_PYR-1 LDREVPGEVLAAIGRDVNAVTLEVVDLT
MAB_3304c|M.abscessus_ATCC_199 IDRTAPDAVLDEIRAAVGATTLELVDLS
MSMEG_2378|M.smegmatis_MC2_155 VDTDVPDDVRAAIADAVGASTLEVVDLS
TH_2037|M.thermoresistible__bu VNRDVPADVRDAIGAAVGASTLEVVDLS
:: .* * * *.* **:*:*: