For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NTDTNSKDREMSIQAPSEKPTASEQPDAGGFAGDTRSDQTFGDAVRSYAQRVRGGDMGSLPAVLGLVVLF VVFGLANDRFLSALNLANLITQAGSICVLAMGLVFVLLLGDIDLSAGVAGGVAACAMALAVVNLGWPWWA AVLVAIACGAVIGLIIGVLRAKLGIPSFVVTLAFFLGLQGVTLKLIGEGGSVRVDNPVIRGLTIANLPVA VGWTIALVVVLAFAGLEIYNYRRKKALQLTHSPLGVVVARAIAVAVVVLGVTFVLSVNRSVNPRVEIRGI PYVLPLILLLLIGMTVLLRRTAYGRHIYAVGGNAEAARRAGISVDRIRVTAFIACSSLAALSGIIAASYA GKVSASSGAGNTLLYAVGAAVIGGTSLFGGKGRALDAVIGGVVVATIANGLGLLNQSSYINFLVTGGVLL LAAGVDAISRRRRSATGL*
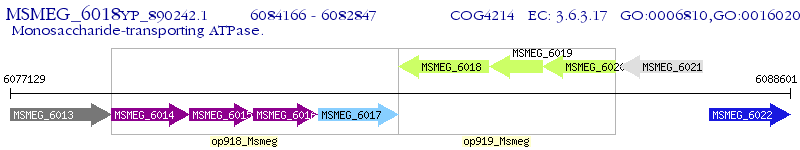
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6018 | - | - | 100% (439) | xylose transport system permease protein XylH |
| M. smegmatis MC2 155 | MSMEG_1706 | - | 6e-49 | 33.42% (386) | xylose transport system permease protein XylH |
| M. smegmatis MC2 155 | MSMEG_4171 | - | 4e-35 | 29.23% (390) | ribose transport system permease protein RbsC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2606 | - | 5e-23 | 25.59% (340) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00397 | - | 2e-18 | 31.82% (154) | putative transporter protein |
| M. abscessus ATCC 19977 | MAB_2624c | - | 2e-05 | 28.93% (121) | high-affinity branched-chain amino acid ABC transporter (LivM) |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1081 | - | 2e-08 | 25.00% (296) | ABC transporter branched chain amino acid transport permease |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5291 | - | 0.0 | 87.99% (408) | inner-membrane translocator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6018|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00397|M.leprae_Br4923 MDHTTKCDAEQYFQAIVTSMADGVIVVDIDGRIESINPAATRILGLRAHD
TH_1081|M.thermoresistible__bu --------------------------------------------------
MAB_2624c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6018|M.smegmatis_MC2_155 -----------------------------MNTDTNSKDREMSIQAPSEK-
Mvan_5291|M.vanbaalenii_PYR-1 -------------------------------MTTVRSDSDVSLADAD---
Mflv_2606|M.gilvum_PYR-GCK ----------------------------------MSTKEALDVGGHK---
MLBr_00397|M.leprae_Br4923 VVDMKHGHPFCFYDTDNQRVDLEREVMRVVRREVTTVSKVVGIDQHSGQR
TH_1081|M.thermoresistible__bu -----------------------------------MTHQCVALYTCH---
MAB_2624c|M.abscessus_ATCC_199 ----------------------------------MSAARALAPGDSIRR-
:
MSMEG_6018|M.smegmatis_MC2_155 -------------------------PTASEQPDAGGFAGDTRSDQTFGDA
Mvan_5291|M.vanbaalenii_PYR-1 ------------------------------------FSGDTRADETFGAA
Mflv_2606|M.gilvum_PYR-GCK ----------------------------------------VVRDERVKER
MLBr_00397|M.leprae_Br4923 LWLSVNVSLLAYKAPPHSALVVSFSDISAHHLSIERLTYEATHDCLTGLA
TH_1081|M.thermoresistible__bu -----------------------------------------AADITFN--
MAB_2624c|M.abscessus_ATCC_199 ------------------------------------WWSELARPVQWVYG
MSMEG_6018|M.smegmatis_MC2_155 VR-------------------SYAQRVRGGDMGSLPAVLGLVVLFVVFGL
Mvan_5291|M.vanbaalenii_PYR-1 VR-------------------GYLQRVRGGDMGSLPAVLGLVVLFVVFGL
Mflv_2606|M.gilvum_PYR-GCK NR-------------------LQRLLIR-PEMGAGIGAIGIFVFFLVVAP
MLBr_00397|M.leprae_Br4923 NRRFAEDQITKSLQHDERSRLAAVLLLDLDDFKVINDSLGHDVGDAVLQT
TH_1081|M.thermoresistible__bu -----------------------LQGLREGLLQLTIDGLSWGAIYALVAV
MAB_2624c|M.abscessus_ATCC_199 VVG-------------------FALLALLPVWKIPYLDTPNTSFGGVMAQ
:.
MSMEG_6018|M.smegmatis_MC2_155 ANDRFLSALNLANLITQAG--SICVLAMG-----------------LVFV
Mvan_5291|M.vanbaalenii_PYR-1 ANDRFLSALNLANLITQAG--SICVLAMG-----------------LVFV
Mflv_2606|M.gilvum_PYR-GCK P---FREASSLATVLYASS--TIGIMACG-----------------VAVL
MLBr_00397|M.leprae_Br4923 VAQRLRSAVRPDDVVARLGGDEFIVLLRGPLSDMNANDVAKRLHTTLSES
TH_1081|M.thermoresistible__bu GYTLVYGVLKLINFAHAEV---FMLGMFG--------------------S
MAB_2624c|M.abscessus_ATCC_199 FAMVALIAIGLNVVVGQAG--LLDLGYVG------------------FYA
. . : : *
MSMEG_6018|M.smegmatis_MC2_155 LLLGDIDLSAGVAGGVAACA------MALAVVNLGWPWWAA---------
Mvan_5291|M.vanbaalenii_PYR-1 LLLGDIDLSAGVAGGVSACA------MALIVVNLSWPWWAA---------
Mflv_2606|M.gilvum_PYR-GCK MIGGEFDLSAGVAVTFSSLA------ASMLAYNLHLNLWVG---------
MLBr_00397|M.leprae_Br4923 LVVDQLTVPIGASVGILEVRPDDRRRAADILRDADSAMYAAKNKKQCAVT
TH_1081|M.thermoresistible__bu YFCLDVILGFTPRGNAYDLG------IGLTVLYLGIAMLFA---------
MAB_2624c|M.abscessus_ATCC_199 IGAYTVALLTSPDSPWNRMG-------TSAFFSSDWAWLSC---------
. :
MSMEG_6018|M.smegmatis_MC2_155 ---VLVAIACGAVIGLIIGVLRAKLGIPS-----FVVTLAFFLGLQGVTL
Mvan_5291|M.vanbaalenii_PYR-1 ---LLVGIACGALIGLAIGLLRAKLGIPS-----FVVTLAFFLGLQGVTL
Mflv_2606|M.gilvum_PYR-GCK ---ALLALVLALTVGFFNGFLVMKTKIPS-----FLITLSTFFMLAGINL
MLBr_00397|M.leprae_Br4923 PQQLVPFVALIALFVFFTAAAGAKFYAPSNLLVILQQTVVLAIVGYGMTF
TH_1081|M.thermoresistible__bu ---IAVSGTAAAGLEFIAYRPLRRRNAAR---------LSFLITAIGMSF
MAB_2624c|M.abscessus_ATCC_199 ---LPIAVAITACFGLILGTPTLRLRGDY---------LAIVTLGFGEII
. . : : : * :
MSMEG_6018|M.smegmatis_MC2_155 KLIGEGGSVRVDNPVIRGLTIANLPVAVGWTIALVVVLAFAGLEIYNYRR
Mvan_5291|M.vanbaalenii_PYR-1 KLIGEGGSVRVDDPVIRGLTISNLPVAAGWAIAGVVVLGFAGLELHQHRR
Mflv_2606|M.gilvum_PYR-GCK AVT---------KLVAGQVATQSVSDMQGWDS------------------
MLBr_00397|M.leprae_Br4923 VIMAGSVDLSVGSIVALTGVTAALVAAQNQFAAIVTALLVGLAAGMVNGI
TH_1081|M.thermoresistible__bu VLQEFVHFILPKILDGYGGSNAQQPIRL----------------------
MAB_2624c|M.abscessus_ATCC_199 RLLADNLDVTNGPRGLHKVAYPHVGEQHVHGGG-----------------
:
MSMEG_6018|M.smegmatis_MC2_155 KKALQLTHSPLGVVVARAIAVAVVVLGVTFVLSVNRSVNPRVEIRGIPYV
Mvan_5291|M.vanbaalenii_PYR-1 KIALRLSHSPLGVVVARVGAVAAVVLGVTYVMSVNRSVSAVADIRGIPYV
Mflv_2606|M.gilvum_PYR-GCK --------------AQKVFASSFTLFG-----------------VGIRIT
MLBr_00397|M.leprae_Br4923 VFAYGKIPSFVSTLGMLQVCRGITLMISDSSAKPMPFHGILGAMGAMPWI
TH_1081|M.thermoresistible__bu ------------VTPETQFTLGDVRVS-----------------NVTLII
MAB_2624c|M.abscessus_ATCC_199 -------------VFSSANSGGHLNYG-----------------VWWYWL
.
MSMEG_6018|M.smegmatis_MC2_155 LPLILLLLIGMTVLLRRTAYGRHIYAVGGNAEAARRAGISVDRIRVTAFI
Mvan_5291|M.vanbaalenii_PYR-1 LPLMLVLLIAMTVVLRRTSYGRHIYAVGGNAEAARRAGISVDRIRVSVFV
Mflv_2606|M.gilvum_PYR-GCK VVWWLVFTAVATWVLFKTKVGNWIFAVGGDADSARAVGVPVTKVKIGLFM
MLBr_00397|M.leprae_Br4923 LIVCLFVTILAGILFQFTMFGRWVKAIGGNERVATLAGVPTRGIKVAIFA
TH_1081|M.thermoresistible__bu VFAALVLALITDVVINRTRLGRGIRAVAQDPDTATLMGVSRERVILTTFV
MAB_2624c|M.abscessus_ATCC_199 GLCLIAAVLVLMGNLERSRVGRSWVAIREDEDAAEMMGVPTFRFKLWAFV
: : : *. *: : * *:. . : *
MSMEG_6018|M.smegmatis_MC2_155 ACSSLAALSGIIAASYAGKVSASSGAGNTLLYAVGAAVIGGTSLFGGKGR
Mvan_5291|M.vanbaalenii_PYR-1 VCSSLAALSGIIAASYAGKVSASSGAGNTLLYAVGAAVIGGTSLFGGKGR
Mflv_2606|M.gilvum_PYR-GCK FVGFCAWFVGMHLLFAFNTVQSGQGIGNEFFYII-AAVIGGCLLTGGYGT
MLBr_00397|M.leprae_Br4923 ICGLTAGLGGIVLASRLGAGTPTAATG-FEIDVIAAVVIGGTPLTGGLGR
TH_1081|M.thermoresistible__bu IGGLLAGAAALLYTLKMPQAIIYSGGFLLGIKAFTAAVLG------GIGN
MAB_2624c|M.abscessus_ATCC_199 IGASIGGLSGALYAGQVQYVASPTFNIINSMLFLCAVVLG------GQGN
. . . : . *.*:* * *
MSMEG_6018|M.smegmatis_MC2_155 ALDAVIGGVVVATIANG-------LGLLNQSSYINFLVTGGVLLLAAGVD
Mvan_5291|M.vanbaalenii_PYR-1 AIDAVIGGVVVATIANG-------LGLLNQSSYINFLVTGGVLLLAASVD
Mflv_2606|M.gilvum_PYR-GCK AIGAAIGALIFGMTNQG-------IVYAGWDPDWFKFFLGAMLLFAVIAN
MLBr_00397|M.leprae_Br4923 LSGTLIGAIIISMLSNG-------MVFMGVGNAASQIIKGIMLAAAVFVF
TH_1081|M.thermoresistible__bu LRGALLGGLLLGVMENYGQ-----AVFGTQWRDVVAFVLLVLVLFFRPTG
MAB_2624c|M.abscessus_ATCC_199 KLGVVVGAFIIVYLPNRLLGVQVQGIDLGNLKYLFFGLALVVLMLFRPQG
.. :*..:. : . ::
MSMEG_6018|M.smegmatis_MC2_155 AISRRRRSATGL-------------------
Mvan_5291|M.vanbaalenii_PYR-1 AISRRRRSSTGLG------------------
Mflv_2606|M.gilvum_PYR-GCK NAFRSYAARK---------------------
MLBr_00397|M.leprae_Br4923 LQRRKIGIIK---------------------
TH_1081|M.thermoresistible__bu ILGESLGKARV--------------------
MAB_2624c|M.abscessus_ATCC_199 LFPAHQKLMAYWRKTSERLSAQKPDEVEAGA