For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSTVNPQSAQADSAPSPANSPATPPTPPGLRTRLSAYLMSHVRQSGMVVALVAIVLLFQVWTSGILLKP LNVTNIIQQNGYILVLAIGMVIVIINGHIDLSVGSVAGFIGAMSAVLMVRNDMPWPVAVVVCVLAGAVIG AWQGFWIAFVGIPSFIVTLGGMLVFRGATQYLLEGQSIAPMPRGLEQVSSGFLPEWGTNALYHWPTVILG AVIMVAAVVVQFRRRRSQIRYGQDVSGVPVFIAKCAAIVVALGAVTLLLASYRGVPIVGIILVVLTLVYA FMMRNTVFGRQVYAVGGNPAAAALSGVRNRWVTLAVFVNMGMLAAIAGLLFAARLNSATPQAGINFELEA IAAAFIGGASSSGGVGTVFGAIVGGLVLGVLNNGMSILGIGSDIQQVIKGLVLLAAVGFDVYNKRKGGR
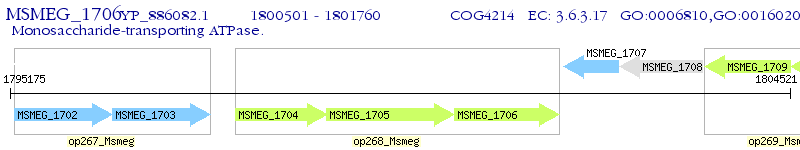
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1706 | - | - | 100% (419) | xylose transport system permease protein XylH |
| M. smegmatis MC2 155 | MSMEG_6018 | - | 6e-49 | 33.42% (386) | xylose transport system permease protein XylH |
| M. smegmatis MC2 155 | MSMEG_3601 | - | 3e-41 | 28.85% (416) | ribose/xylose/arabinose/galactoside ABC-type transport systems, |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2606 | - | 1e-19 | 23.14% (389) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00397 | - | 1e-30 | 28.46% (369) | putative transporter protein |
| M. abscessus ATCC 19977 | MAB_2625c | - | 4e-12 | 33.59% (128) | branched chain amino acid ABC transporter |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1081 | - | 8e-12 | 34.40% (125) | ABC transporter branched chain amino acid transport permease |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5291 | - | 1e-47 | 32.38% (386) | inner-membrane translocator |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1706|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 MDHTTKCDAEQYFQAIVTSMADGVIVVDIDGRIESINPAATRILGLRAHD
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1706|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 VVDMKHGHPFCFYDTDNQRVDLEREVMRVVRREVTTVSKVVGIDQHSGQR
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 -----------------------MISGWMAQTGVQASTISFD--------
TH_1081|M.thermoresistible__bu -----------------------MTHQCVALYTCHAADITFN--------
Mvan_5291|M.vanbaalenii_PYR-1 ------------------------MTTVRSDSDVSLADADFSGDTRADET
MSMEG_1706|M.smegmatis_MC2_155 ------------------------MTSTVNPQSAQADSAPSPANSPATPP
MLBr_00397|M.leprae_Br4923 LWLSVNVSLLAYKAPPHSALVVSFSDISAHHLSIERLTYEATHDCLTGLA
Mflv_2606|M.gilvum_PYR-GCK -----------------------MSTKEALDVGGHKVVRDERVKE-----
MAB_2625c|M.abscessus_ATCC_199 --------LHGLWSSIGQLTIDGLAWGAIYALVAVGYT---------LVF
TH_1081|M.thermoresistible__bu --------LQGLREGLLQLTIDGLSWGAIYALVAVGYT---------LVY
Mvan_5291|M.vanbaalenii_PYR-1 FGAAVRGYLQRVRGGDMGSLPAVLGLVVLFVVFGLAND---------RFL
MSMEG_1706|M.smegmatis_MC2_155 TPPGLRTRLSAYLMSHVRQSGMVVALVAIVLLFQVWTSG--------ILL
MLBr_00397|M.leprae_Br4923 NRRFAEDQITKSLQHDERSRLAAVLLLDLDDFKVINDSLGHDVGDAVLQT
Mflv_2606|M.gilvum_PYR-GCK -----RNRLQRLLIR--PEMGAGIGAIGIFVFFLVVAP---------PFR
: : : . :
MAB_2625c|M.abscessus_ATCC_199 GVLRLINFAHSE----VFMLGMFGAYFAL------------------DVV
TH_1081|M.thermoresistible__bu GVLKLINFAHAE----VFMLGMFGSYFCL------------------DVI
Mvan_5291|M.vanbaalenii_PYR-1 SALNLANLITQAGSICVLAMGLVFVLLLG------------------DID
MSMEG_1706|M.smegmatis_MC2_155 KPLNVTNIIQQNGYILVLAIGMVIVIING------------------HID
MLBr_00397|M.leprae_Br4923 VAQRLRSAVRPDDVVARLGGDEFIVLLRGPLSDMNANDVAKRLHTTLSES
Mflv_2606|M.gilvum_PYR-GCK EASSLATVLYASSTIGIMACGVAVLMIGG------------------EFD
: . : . :
MAB_2625c|M.abscessus_ATCC_199 LGFSPGGNAYNKGVSLTVLYLG----VAMLVAMAVSATTALG--LEAVAY
TH_1081|M.thermoresistible__bu LGFTPRGNAYDLGIGLTVLYLG----IAMLFAIAVSGTAAAG--LEFIAY
Mvan_5291|M.vanbaalenii_PYR-1 LSAGVAGGVSACAMALIVVNLSWPWWAALLVGIACGALIGLA--IGLLRA
MSMEG_1706|M.smegmatis_MC2_155 LSVGSVAGFIGAMSAVLMVRNDMPWPVAVVVCVLAGAVIGAW--QGFWIA
MLBr_00397|M.leprae_Br4923 LVVDQLTVPIGASVGILEVRPDDRRRAADILRDADSAMYAAKNKKQCAVT
Mflv_2606|M.gilvum_PYR-GCK LSAGVAVTFSSLAASMLAYNLHLNLWVGALLALVLALTVGFFN--GFLVM
* .: . :. . .
MAB_2625c|M.abscessus_ATCC_199 RPLRRRGARPLTFLITAIGMS-----------------------------
TH_1081|M.thermoresistible__bu RPLRRRNAARLSFLITAIGMS-----------------------------
Mvan_5291|M.vanbaalenii_PYR-1 KLGIPSFVVTLAFFLGLQGVTLKLIG-------EGGSVRVDDPVIRGLTI
MSMEG_1706|M.smegmatis_MC2_155 FVGIPSFIVTLGGMLVFRGATQYLLEGQSIAPMPRGLEQVSSGFLPEWGT
MLBr_00397|M.leprae_Br4923 PQQLVPFVALIALFVFFTAAAGAKFYAPSNLLVILQQTVVLAIVGYGMTF
Mflv_2606|M.gilvum_PYR-GCK KTKIPSFLITLSTFFMLAGIN-----------------------------
: :. .
MAB_2625c|M.abscessus_ATCC_199 -------------FVIQEFVHWVLPKIAKGYGGSNAQQPLMLVTPRPVWT
TH_1081|M.thermoresistible__bu -------------FVLQEFVHFILPKILDGYGGSNAQQPIRLVTPETQFT
Mvan_5291|M.vanbaalenii_PYR-1 SNLPVAAGWAIAGVVVLGFAGLELHQHRRKIALRLSHSPLGVVVARVGAV
MSMEG_1706|M.smegmatis_MC2_155 NALYHWPTVILGAVIMVAAVVVQFRRRRSQIRYGQDVSGVPVFIAKCAAI
MLBr_00397|M.leprae_Br4923 VIMAGSVDLSVGSIVALTGVTAALVAAQNQFAAIVTALLVGLAAGMVNGI
Mflv_2606|M.gilvum_PYR-GCK --------------LAVTKLVAGQVATQSVSDMQGWDSAQKVFASSFTLF
: :
MAB_2625c|M.abscessus_ATCC_199 FGPFDVSGHHIPFQATITNVTVV--------------------------V
TH_1081|M.thermoresistible__bu LG--DVR---------VSNVTLI--------------------------I
Mvan_5291|M.vanbaalenii_PYR-1 AAVVLGVTYVMSVNRSVSAVADIRGIP---------------------YV
MSMEG_1706|M.smegmatis_MC2_155 VVALGAVTLLLASYRGVPIVGII---------------------------
MLBr_00397|M.leprae_Br4923 VFAYGKIPSFVSTLGMLQVCRGITLMISDSSAKPMPFHGILGAMGAMPWI
Mflv_2606|M.gilvum_PYR-GCK GVGIRIT-------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 VAAALVLAFVTDIAINHTKFGRGIRAVAQDSTTATLMGVSRERIIMATFV
TH_1081|M.thermoresistible__bu VFAALVLALITDVVINRTRLGRGIRAVAQDPDTATLMGVSRERVILTTFV
Mvan_5291|M.vanbaalenii_PYR-1 LPLMLVLLIAMTVVLRRTSYGRHIYAVGGNAEAARRAGISVDRIRVSVFV
MSMEG_1706|M.smegmatis_MC2_155 ---LVVLTLVYAFMMRNTVFGRQVYAVGGNPAAAALSGVRNRWVTLAVFV
MLBr_00397|M.leprae_Br4923 LIVCLFVTILAGILFQFTMFGRWVKAIGGNERVATLAGVPTRGIKVAIFA
Mflv_2606|M.gilvum_PYR-GCK VVWWLVFTAVATWVLFKTKVGNWIFAVGGDADSARAVGVPVTKVKIGLFM
:.. : * *. : *:. : * *: : : *
MAB_2625c|M.abscessus_ATCC_199 IGGLLAGAAALLYTLKVPQGIIYSGGFLLGIKAFSAAVLGGIGNLRG---
TH_1081|M.thermoresistible__bu IGGLLAGAAALLYTLKMPQAIIYSGGFLLGIKAFTAAVLGGIGNLRG---
Mvan_5291|M.vanbaalenii_PYR-1 VCSSLAALSGIIAASYAGKVSASSGAGNTLLYAVGAAVIGGTSLFGGKGR
MSMEG_1706|M.smegmatis_MC2_155 NMGMLAAIAGLLFAARLNSATPQAG-INFELEAIAAAFIGGASSSGGVGT
MLBr_00397|M.leprae_Br4923 ICGLTAGLGGIVLASRLGAGTPTAA-TGFEIDVIAAVVIGGTPLTGGLGR
Mflv_2606|M.gilvum_PYR-GCK FVGFCAWFVGMHLLFAFNTVQSGQG-IGNEFFYIIAAVIGGCLLTGGYGT
. * .: . : . *..:** *
MAB_2625c|M.abscessus_ATCC_199 ---ALLGGLILGVMENYGQAVFGTQWRDVVAFVLLVLVLMFRPTGILGES
TH_1081|M.thermoresistible__bu ---ALLGGLLLGVMENYGQAVFGTQWRDVVAFVLLVLVLFFRPTGILGES
Mvan_5291|M.vanbaalenii_PYR-1 AIDAVIGGVVVATIAN-GLGLLNQSSYINFLVTGGVLLLAASVDAISRRR
MSMEG_1706|M.smegmatis_MC2_155 VFGAIVGGLVLGVLNN-GMSILGIGSDIQQVIKGLVLLAAVGFDVYNKRK
MLBr_00397|M.leprae_Br4923 LSGTLIGAIIISMLSN-GMVFMGVGNAASQIIKG-IMLAAAVFVFLQRRK
Mflv_2606|M.gilvum_PYR-GCK AIGAAIGALIFGMTNQ-GIVYAGWDPDWFKFFLGAMLLFAVIANNAFRSY
: :*.::.. : * . . :::
MAB_2625c|M.abscessus_ATCC_199 LGRAKA-
TH_1081|M.thermoresistible__bu LGKARV-
Mvan_5291|M.vanbaalenii_PYR-1 RSSTGLG
MSMEG_1706|M.smegmatis_MC2_155 GGR----
MLBr_00397|M.leprae_Br4923 IGIIK--
Mflv_2606|M.gilvum_PYR-GCK AARK---
.