For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRLMTPLTELVGIEHPVVQTGMGWVAGARLVSATSNAGGLGILASATMTLDELKTAVAKVKAATDKPFG INIRADAGDANERVDLLISEGVKVASFALAPKPDLIAKLKEAGVVVIPSVGLAKHAKKVASWGADAVIVQ GGEGGGHTGPIATTLLLPSVLDAVADTGMPVIAAGGFFDGRGLAAALSYGAAGVAMGTRFLLTSDSTVPD AVKQRYLQAALDGTVVSTRVDGMPHRVLRTGLVEKLESGSRVRGFAAAVGNAQKFKKMSGMTWRSMIKDG LAMRHGKELTWSQVVMAANTPMLLKAGLVEGNTDAGVLASGQVTGIIDDLPSCAELVPAIVADAVEHLNK ASSFIQEK
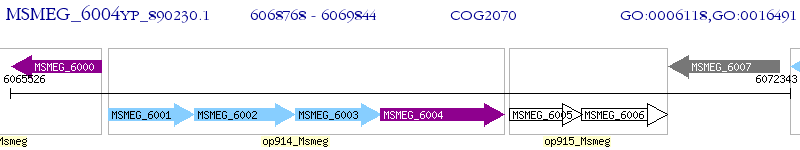
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6004 | - | - | 100% (358) | oxidoreductase, 2-nitropropane dioxygenase family protein |
| M. smegmatis MC2 155 | MSMEG_5182 | - | 5e-38 | 30.97% (352) | 2-nitropropane dioxygenase |
| M. smegmatis MC2 155 | MSMEG_1660 | - | 5e-27 | 34.02% (241) | oxidoreductase, 2-nitropropane dioxygenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3583 | - | 1e-169 | 83.05% (354) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1487 | - | 0.0 | 92.13% (356) | 2-nitropropane dioxygenase, NPD |
| M. tuberculosis H37Rv | Rv3553 | - | 1e-169 | 83.05% (354) | oxidoreductase |
| M. leprae Br4923 | MLBr_00387 | guaB2 | 1e-05 | 31.94% (144) | inosine 5'-monophosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0603c | - | 1e-168 | 84.94% (352) | 2-nitropropane dioxygenase |
| M. marinum M | MMAR_5042 | - | 1e-171 | 83.90% (354) | 2-nitropropane dioxygenase |
| M. avium 104 | MAV_0607 | - | 1e-173 | 85.63% (355) | oxidoreductase, 2-nitropropane dioxygenase family protein |
| M. thermoresistible (build 8) | TH_1758 | - | 1e-176 | 85.99% (357) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4116 | - | 1e-171 | 83.62% (354) | 2-nitropropane dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_5269 | - | 0.0 | 92.98% (356) | 2-nitropropane dioxygenase, NPD |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3583|M.bovis_AF2122/97 --------------------------------------------------
Rv3553|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5042|M.marinum_M --------------------------------------------------
MUL_4116|M.ulcerans_Agy99 --------------------------------------------------
MAV_0607|M.avium_104 --------------------------------------------------
Mflv_1487|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5269|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6004|M.smegmatis_MC2_155 --------------------------------------------------
TH_1758|M.thermoresistible__bu --------------------------------------------------
MAB_0603c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00387|M.leprae_Br4923 MIRGMSNLKESSDFVASSYVRLGGLMDDPAATGGDNPHKVAMLGLTFDDV
Mb3583|M.bovis_AF2122/97 -------------MRLRTPLTELIGIEHPVVQTGMGWVAGARLVSATANA
Rv3553|M.tuberculosis_H37Rv -------------MRLRTPLTELIGIEHPVVQTGMGWVAGARLVSATANA
MMAR_5042|M.marinum_M -------------MRLRTPLTDLVGVEHPVVQTGMGWVAGARLVSATANA
MUL_4116|M.ulcerans_Agy99 -------------MRLRTPLTDLVGVEHPVVQTGMGWVAGARLVSATANA
MAV_0607|M.avium_104 -----------MDMRLRTPLTELVGVEHPVVQTGMGWVAGARLVSATANA
Mflv_1487|M.gilvum_PYR-GCK ------------MSTLKTPLTELVGIEHPVVQTGMGWVAGARLVAATSNA
Mvan_5269|M.vanbaalenii_PYR-1 ------------MSRLSTPLTELVGIEHPVVQTGMGWVAGARLVSATSNA
MSMEG_6004|M.smegmatis_MC2_155 ------------MSRLMTPLTELVGIEHPVVQTGMGWVAGARLVSATSNA
TH_1758|M.thermoresistible__bu ------------LSRIRTPLTDLVGIEHPVVQTGMGWVAGARLVAATANA
MAB_0603c|M.abscessus_ATCC_199 -----------MTQVLKTALTQLVGIEHPVVQTGMGWVAGPRLVAATSNA
MLBr_00387|M.leprae_Br4923 LLLPAASDVVPATADISSQLTKKIRLKVPLVSSAMDTVTEARMAIAMARA
: : **. : :: *:*.:.*. *: .*:. * :.*
Mb3583|M.bovis_AF2122/97 GGLGILAS-----------ATMTLDELAAAITKVKAVTDKPFGVN-----
Rv3553|M.tuberculosis_H37Rv GGLGILAS-----------ATMTLDELAAAITKVKAVTDKPFGVN-----
MMAR_5042|M.marinum_M GGLGILAS-----------ATMTLDELATAIGKVKATTDKPFGVN-----
MUL_4116|M.ulcerans_Agy99 GGLGILAS-----------ATMTLDELATAIGKVKATTDKPFGVN-----
MAV_0607|M.avium_104 GGLGILAS-----------ATMTLDELATAIAKVKAATDKPFGVN-----
Mflv_1487|M.gilvum_PYR-GCK GGLGILAS-----------ATMTLTELTTAVAKVKAATDKPFGIN-----
Mvan_5269|M.vanbaalenii_PYR-1 GGLGILAS-----------ATMTLEELTTAVAKVKAATDKPFGIN-----
MSMEG_6004|M.smegmatis_MC2_155 GGLGILAS-----------ATMTLDELKTAVAKVKAATDKPFGIN-----
TH_1758|M.thermoresistible__bu GGLGILAS-----------ATMTLDELQTAVDKVKAATDKPFGIN-----
MAB_0603c|M.abscessus_ATCC_199 GGLGILAS-----------ATMTLDELIAAVAKTKSLTDKPFGVN-----
MLBr_00387|M.leprae_Br4923 GGMGVLHRNLPVGEQAGQVETVKRSEAGMVTDPVTCRPDNTLAQVGALCA
**:*:* *:. * . ... .*:.:.
Mb3583|M.bovis_AF2122/97 ------------------------IRADAADAGDRVELMIREGVRVASFA
Rv3553|M.tuberculosis_H37Rv ------------------------IRADAADAGDRVELMIREGVRVASFA
MMAR_5042|M.marinum_M ------------------------IRADAADASDRVELMIREGVKVASFA
MUL_4116|M.ulcerans_Agy99 ------------------------IRADAADASDRVELMIREGVKVASFA
MAV_0607|M.avium_104 ------------------------IRADAADAADRVDLMIREGVKVASFA
Mflv_1487|M.gilvum_PYR-GCK ------------------------IRADAGDAGERVDLLIREGVKVASFA
Mvan_5269|M.vanbaalenii_PYR-1 ------------------------IRADAGDANARVDLLIREGVKVASFA
MSMEG_6004|M.smegmatis_MC2_155 ------------------------IRADAGDANERVDLLISEGVKVASFA
TH_1758|M.thermoresistible__bu ------------------------IRADAGDANERVDLLIREGVKVASFA
MAB_0603c|M.abscessus_ATCC_199 ------------------------IRADAGDATERIDLLIREKVKVASFA
MLBr_00387|M.leprae_Br4923 RFRISGLPVVDDSGALAGIITNRDMRFEVDQSKQVAEVMTKTPLITAAEG
:* :. :: ::: : .*: .
Mb3583|M.bovis_AF2122/97 LAPKQQLIARLKE--------------AGAVVIPSIGAAKHARKVAAWGA
Rv3553|M.tuberculosis_H37Rv LAPKQQLIARLKE--------------AGAVVIPSIGAAKHARKVAAWGA
MMAR_5042|M.marinum_M LAPKPELIARLKE--------------AGAVVIPSIGAAKHARKVAGWGA
MUL_4116|M.ulcerans_Agy99 LAPKPELIARLKE--------------AGAVVIPSIGAAKHARKVAGWGA
MAV_0607|M.avium_104 LAPKQELIARLKE--------------AGAVVIPSIGAAKHARKVAGWGA
Mflv_1487|M.gilvum_PYR-GCK LAPKPDLIARLKE--------------AGVVVIPSVGLAKHAKKVAGWGA
Mvan_5269|M.vanbaalenii_PYR-1 LAPKPDLIARLKD--------------AGVVVIPSVGLAKHAKKVAGWGA
MSMEG_6004|M.smegmatis_MC2_155 LAPKPDLIAKLKE--------------AGVVVIPSVGLAKHAKKVASWGA
TH_1758|M.thermoresistible__bu LAPKPELIARLKD--------------AGVVVIPSVGAARHAEKVAGWGA
MAB_0603c|M.abscessus_ATCC_199 LAPKQDLIAKLKD--------------NGVVVVPSIGAAKHAKKVAAWGA
MLBr_00387|M.leprae_Br4923 VSADAALGLLRRNKIEKLPVVDGHGRLTGLITVKDFVKTEQHPLATKDND
::.. * :: * :.: .. :.: .: .
Mb3583|M.bovis_AF2122/97 DAMIVQGGEGGG--------------------HTGPVATTLLLPSVLDAV
Rv3553|M.tuberculosis_H37Rv DAMIVQGGEGGG--------------------HTGPVATTLLLPSVLDAV
MMAR_5042|M.marinum_M DAMIVQGGEGGG--------------------HTGPVATTLLLPSVLDAV
MUL_4116|M.ulcerans_Agy99 DAMIVQGGEGGG--------------------HTGPVATTLLLPSVLDAV
MAV_0607|M.avium_104 DAMIVQGGEGGG--------------------HTGPVATTLLLPSVLDAV
Mflv_1487|M.gilvum_PYR-GCK DAVIVQGGEGGG--------------------HTGPIATTLLLPSVLDAV
Mvan_5269|M.vanbaalenii_PYR-1 DAVIVQGGEGGG--------------------HTGPIATTLLLPSVLDAV
MSMEG_6004|M.smegmatis_MC2_155 DAVIVQGGEGGG--------------------HTGPIATTLLLPSVLDAV
TH_1758|M.thermoresistible__bu DAVIVQGGEGGG--------------------HTGPVPTTLLLPSVLDAV
MAB_0603c|M.abscessus_ATCC_199 DAVIVQGGEGGG--------------------HTGPVATTLLLPSVLDAV
MLBr_00387|M.leprae_Br4923 GRLLVGAAVGVGGDAWVRAMMLVDAGVDVLIVDTAHAHNRLVLDMVGKLK
. ::* .. * * .*. . *:* * .
Mb3583|M.bovis_AF2122/97 AGTGIPVIAAGGFFDGRGLAAALCYG-AAGVAMGTRFLLTSDSTVPDAVK
Rv3553|M.tuberculosis_H37Rv AGTGIPVIAAGGFFDGRGLAAALCYG-AAGVAMGTRFLLTSDSTVPDAVK
MMAR_5042|M.marinum_M KGTGIPVIAAGGFFDGRGLAAALSYG-AAGVAMGTRFLLTSDSTVPEAVK
MUL_4116|M.ulcerans_Agy99 KGTGIPVIAAGGFFDGHGLAAALSYG-AAGVAMGTRFLLTSDSTVPDAVK
MAV_0607|M.avium_104 RGTGIPVIAAGGFFDGRGLAAALSYG-AAGVAMGTRFLLTSDSTVPDAVK
Mflv_1487|M.gilvum_PYR-GCK ADTGMPVIAAGGFFDGRGLAAALTYG-AAGVAMGTRFLLTKDSTVPDAVK
Mvan_5269|M.vanbaalenii_PYR-1 ADTGMPVIAAGGFFDGRGLAAALSYG-AAGVAMGTRFLLTSDSTVPDAVK
MSMEG_6004|M.smegmatis_MC2_155 ADTGMPVIAAGGFFDGRGLAAALSYG-AAGVAMGTRFLLTSDSTVPDAVK
TH_1758|M.thermoresistible__bu RDTGMPVVAAGGFFDGRGLAAALAYG-AAGVAMGTRFLLTKDSTVPDAVK
MAB_0603c|M.abscessus_ATCC_199 KDTGMPVVAAGGFFDGRGLAAALSYG-AAGVAMGTRFLLTSDSSVPDGVK
MLBr_00387|M.leprae_Br4923 VEIGDRVQVIGGNVATRSAAAALVEAGADAVKVGVGPGSTCTTRVVAGVG
* * . ** . :. **** . * .* :*. * : * .*
Mb3583|M.bovis_AF2122/97 RRYLQAGLDGTVVT--TRVDGMPHRVLRTE--LVEKLES-------GSRA
Rv3553|M.tuberculosis_H37Rv RRYLQAGLDGTVVT--TRVDGMPHRVLRTE--LVEKLES-------GSRA
MMAR_5042|M.marinum_M RRYLQAALDGTVVT--TRVDGMPHRVLRTG--LVEKLEG-------GSRA
MUL_4116|M.ulcerans_Agy99 RRYLQAALDGTVVT--TRVDGMPHRVLRTG--LVEKLEG-------GSRA
MAV_0607|M.avium_104 KRYLEAALDGTVVT--TRVDGMPHRVLRTG--LVEKLES-------GSPV
Mflv_1487|M.gilvum_PYR-GCK QRYLDAALDGTVVS--TKVDGMPHRVLRTG--LVEKLES-------GSPV
Mvan_5269|M.vanbaalenii_PYR-1 QRYLDAALDGTVVS--TKVDGMPHRVLRTG--LVEKLES-------GSPI
MSMEG_6004|M.smegmatis_MC2_155 QRYLQAALDGTVVS--TRVDGMPHRVLRTG--LVEKLES-------GSRV
TH_1758|M.thermoresistible__bu RRYLEAGLDGTVVS--TRVDGMPHRVLRTG--LVEKLES-------GSPI
MAB_0603c|M.abscessus_ATCC_199 QRYLASDLAGTVVS--TRVDGMPHRVLRTE--LVEKLES-------GSRV
MLBr_00387|M.leprae_Br4923 APQITAILEAVAACGPAGVPVIADGGLQYSGDIAKALAAGASTTMLGSLL
: : * .... : * :.. *: :.: * . **
Mb3583|M.bovis_AF2122/97 RGFAAALRNAGKFRRMSQMTWRSMIRDGLTMRHGKELTWSQVLMAANTPM
Rv3553|M.tuberculosis_H37Rv RGFAAALRNAGKFRRMSQMTWRSMIRDGLTMRHGKELTWSQVLMAANTPM
MMAR_5042|M.marinum_M RGFTAAIRNAAKFKQMSQMSWASMIKDGLAMRHGKELTWSQVVMAANTPM
MUL_4116|M.ulcerans_Agy99 RGFTAAIRNAAKFKQMSQMSWASMIKDGLAMRHGKELTWSQVLMAANTPM
MAV_0607|M.avium_104 RGLTAAVRNAAKFKHMSKMTWRSMISDGLAMRHGKEMTWSQVVMAANTPM
Mflv_1487|M.gilvum_PYR-GCK RGLSAAVLNAQKFKKMSGMTWKSMITDGLAMRHGKDLTWSQVVMAANTPM
Mvan_5269|M.vanbaalenii_PYR-1 RGLSAAVLNAQKFKKMSGMTWKSMITDGLAMRHGKDLTWSQVVMAANTPM
MSMEG_6004|M.smegmatis_MC2_155 RGFAAAVGNAQKFKKMSGMTWRSMIKDGLAMRHGKELTWSQVVMAANTPM
TH_1758|M.thermoresistible__bu RGFTAAVRNAQKFKKMTGMTWRSMIRDGLEMRRGKDLTWAQVIMAANTPM
MAB_0603c|M.abscessus_ATCC_199 RGFTAAVRNAAKFKKMTGMSWASMVKDGLAMRHGKELTWSQVLMAANTPM
MLBr_00387|M.leprae_Br4923 AGTAEAPGELIFVNGKQFKSYRGMGSLGAMQGRGGDKSYSKDRYFADDAL
* : * : .. :: .* * :* : :::: *: .:
Mb3583|M.bovis_AF2122/97 LLKAGLVDGNTEAGVLASGQVAGILDDLPSCKELIESIVLDAITHLQTAS
Rv3553|M.tuberculosis_H37Rv LLKAGLVDGNTEAGVLASGQVAGILDDLPSCKELIESIVLDAITHLQTAS
MMAR_5042|M.marinum_M LLKAGLVDGNPEAGVLASGQVAGILDDLPSCAQLVESIVHDAIEHLRAAA
MUL_4116|M.ulcerans_Agy99 LLKAGLVDGNPEAGVLASGQVAGILDDLPSCAQLVESIVHDAIEHLRAAA
MAV_0607|M.avium_104 LLKAGLVEGNTEAGVLASGQVAGILEDLPSCAELIDSIVRDAIAHLRSAS
Mflv_1487|M.gilvum_PYR-GCK LLKAGLVEGNTEAGVLASGQVAGIIDDLPSCAELVPAIVAEAVEHLQKAS
Mvan_5269|M.vanbaalenii_PYR-1 LLKAGLVEGNTDAGVLASGQVAGIIDDLPSCAELVPAIVAEAVEHLQKAS
MSMEG_6004|M.smegmatis_MC2_155 LLKAGLVEGNTDAGVLASGQVTGIIDDLPSCAELVPAIVADAVEHLNKAS
TH_1758|M.thermoresistible__bu LLKAGLVEGNTEAGVLASGQVAGILDDLPSCAELIEQIVADAVRHLKGAV
MAB_0603c|M.abscessus_ATCC_199 LLKAGLVEGNTNAGVLASGQVAGILDDLPSCHELIDAIVTDAVAHLKAAN
MLBr_00387|M.leprae_Br4923 SEDK-LVPEGIEGRVPFRGPLSSVIHQLVGGLRAAMGYTGSPTIEVLQQA
. ** . :. * * ::.::.:* . . . .. .:
Mb3583|M.bovis_AF2122/97 ALVE--------------------------
Rv3553|M.tuberculosis_H37Rv ALVE--------------------------
MMAR_5042|M.marinum_M ALVE--------------------------
MUL_4116|M.ulcerans_Agy99 ALVE--------------------------
MAV_0607|M.avium_104 GLIEE-------------------------
Mflv_1487|M.gilvum_PYR-GCK QYIR--------------------------
Mvan_5269|M.vanbaalenii_PYR-1 QHIR--------------------------
MSMEG_6004|M.smegmatis_MC2_155 SFIQEK------------------------
TH_1758|M.thermoresistible__bu ALVEE-------------------------
MAB_0603c|M.abscessus_ATCC_199 ATIL--------------------------
MLBr_00387|M.leprae_Br4923 QFVRITPAGLKESHPHDVAMTVEAPNYYPR
: