For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQVLKTALTQLVGIEHPVVQTGMGWVAGPRLVAATSNAGGLGILASATMTLDELIAAVAKTKSLTDKPFG VNIRADAGDATERIDLLIREKVKVASFALAPKQDLIAKLKDNGVVVVPSIGAAKHAKKVAAWGADAVIVQ GGEGGGHTGPVATTLLLPSVLDAVKDTGMPVVAAGGFFDGRGLAAALSYGAAGVAMGTRFLLTSDSSVPD GVKQRYLASDLAGTVVSTRVDGMPHRVLRTELVEKLESGSRVRGFTAAVRNAAKFKKMTGMSWASMVKDG LAMRHGKELTWSQVLMAANTPMLLKAGLVEGNTNAGVLASGQVAGILDDLPSCHELIDAIVTDAVAHLKA ANATIL*
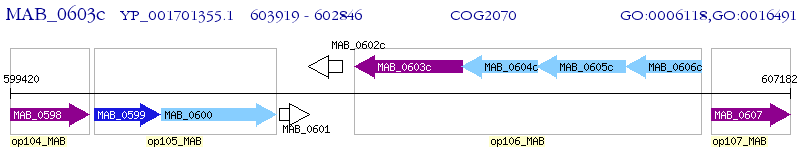
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0603c | - | - | 100% (357) | 2-nitropropane dioxygenase |
| M. abscessus ATCC 19977 | MAB_4001c | - | 1e-34 | 34.41% (247) | putative 2-nitropropane dioxygenase, NPD |
| M. abscessus ATCC 19977 | MAB_4162c | - | 7e-26 | 30.64% (297) | putative 2-nitropropane dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3583 | - | 1e-164 | 80.68% (352) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1487 | - | 1e-165 | 83.24% (352) | 2-nitropropane dioxygenase, NPD |
| M. tuberculosis H37Rv | Rv3553 | - | 1e-164 | 80.68% (352) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5042 | - | 1e-164 | 80.97% (352) | 2-nitropropane dioxygenase |
| M. avium 104 | MAV_0607 | - | 1e-165 | 82.10% (352) | oxidoreductase, 2-nitropropane dioxygenase family protein |
| M. smegmatis MC2 155 | MSMEG_6004 | - | 1e-168 | 84.94% (352) | oxidoreductase, 2-nitropropane dioxygenase family protein |
| M. thermoresistible (build 8) | TH_1758 | - | 1e-166 | 83.24% (352) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4116 | - | 1e-164 | 81.25% (352) | 2-nitropropane dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_5269 | - | 1e-165 | 82.95% (352) | 2-nitropropane dioxygenase, NPD |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3583|M.bovis_AF2122/97 --MRLRTPLTELIGIEHPVVQTGMGWVAGARLVSATANAGGLGILASATM
Rv3553|M.tuberculosis_H37Rv --MRLRTPLTELIGIEHPVVQTGMGWVAGARLVSATANAGGLGILASATM
MMAR_5042|M.marinum_M --MRLRTPLTDLVGVEHPVVQTGMGWVAGARLVSATANAGGLGILASATM
MUL_4116|M.ulcerans_Agy99 --MRLRTPLTDLVGVEHPVVQTGMGWVAGARLVSATANAGGLGILASATM
MAV_0607|M.avium_104 MDMRLRTPLTELVGVEHPVVQTGMGWVAGARLVSATANAGGLGILASATM
TH_1758|M.thermoresistible__bu -LSRIRTPLTDLVGIEHPVVQTGMGWVAGARLVAATANAGGLGILASATM
Mflv_1487|M.gilvum_PYR-GCK -MSTLKTPLTELVGIEHPVVQTGMGWVAGARLVAATSNAGGLGILASATM
Mvan_5269|M.vanbaalenii_PYR-1 -MSRLSTPLTELVGIEHPVVQTGMGWVAGARLVSATSNAGGLGILASATM
MSMEG_6004|M.smegmatis_MC2_155 -MSRLMTPLTELVGIEHPVVQTGMGWVAGARLVSATSNAGGLGILASATM
MAB_0603c|M.abscessus_ATCC_199 MTQVLKTALTQLVGIEHPVVQTGMGWVAGPRLVAATSNAGGLGILASATM
: *.**:*:*:**************.***:**:*************
Mb3583|M.bovis_AF2122/97 TLDELAAAITKVKAVTDKPFGVNIRADAADAGDRVELMIREGVRVASFAL
Rv3553|M.tuberculosis_H37Rv TLDELAAAITKVKAVTDKPFGVNIRADAADAGDRVELMIREGVRVASFAL
MMAR_5042|M.marinum_M TLDELATAIGKVKATTDKPFGVNIRADAADASDRVELMIREGVKVASFAL
MUL_4116|M.ulcerans_Agy99 TLDELATAIGKVKATTDKPFGVNIRADAADASDRVELMIREGVKVASFAL
MAV_0607|M.avium_104 TLDELATAIAKVKAATDKPFGVNIRADAADAADRVDLMIREGVKVASFAL
TH_1758|M.thermoresistible__bu TLDELQTAVDKVKAATDKPFGINIRADAGDANERVDLLIREGVKVASFAL
Mflv_1487|M.gilvum_PYR-GCK TLTELTTAVAKVKAATDKPFGINIRADAGDAGERVDLLIREGVKVASFAL
Mvan_5269|M.vanbaalenii_PYR-1 TLEELTTAVAKVKAATDKPFGINIRADAGDANARVDLLIREGVKVASFAL
MSMEG_6004|M.smegmatis_MC2_155 TLDELKTAVAKVKAATDKPFGINIRADAGDANERVDLLISEGVKVASFAL
MAB_0603c|M.abscessus_ATCC_199 TLDELIAAVAKTKSLTDKPFGVNIRADAGDATERIDLLIREKVKVASFAL
** ** :*: *.*: ******:******.** *::*:* * *:******
Mb3583|M.bovis_AF2122/97 APKQQLIARLKEAGAVVIPSIGAAKHARKVAAWGADAMIVQGGEGGGHTG
Rv3553|M.tuberculosis_H37Rv APKQQLIARLKEAGAVVIPSIGAAKHARKVAAWGADAMIVQGGEGGGHTG
MMAR_5042|M.marinum_M APKPELIARLKEAGAVVIPSIGAAKHARKVAGWGADAMIVQGGEGGGHTG
MUL_4116|M.ulcerans_Agy99 APKPELIARLKEAGAVVIPSIGAAKHARKVAGWGADAMIVQGGEGGGHTG
MAV_0607|M.avium_104 APKQELIARLKEAGAVVIPSIGAAKHARKVAGWGADAMIVQGGEGGGHTG
TH_1758|M.thermoresistible__bu APKPELIARLKDAGVVVIPSVGAARHAEKVAGWGADAVIVQGGEGGGHTG
Mflv_1487|M.gilvum_PYR-GCK APKPDLIARLKEAGVVVIPSVGLAKHAKKVAGWGADAVIVQGGEGGGHTG
Mvan_5269|M.vanbaalenii_PYR-1 APKPDLIARLKDAGVVVIPSVGLAKHAKKVAGWGADAVIVQGGEGGGHTG
MSMEG_6004|M.smegmatis_MC2_155 APKPDLIAKLKEAGVVVIPSVGLAKHAKKVASWGADAVIVQGGEGGGHTG
MAB_0603c|M.abscessus_ATCC_199 APKQDLIAKLKDNGVVVVPSIGAAKHAKKVAAWGADAVIVQGGEGGGHTG
*** :***:**: *.**:**:* *:**.***.*****:************
Mb3583|M.bovis_AF2122/97 PVATTLLLPSVLDAVAGTGIPVIAAGGFFDGRGLAAALCYGAAGVAMGTR
Rv3553|M.tuberculosis_H37Rv PVATTLLLPSVLDAVAGTGIPVIAAGGFFDGRGLAAALCYGAAGVAMGTR
MMAR_5042|M.marinum_M PVATTLLLPSVLDAVKGTGIPVIAAGGFFDGRGLAAALSYGAAGVAMGTR
MUL_4116|M.ulcerans_Agy99 PVATTLLLPSVLDAVKGTGIPVIAAGGFFDGHGLAAALSYGAAGVAMGTR
MAV_0607|M.avium_104 PVATTLLLPSVLDAVRGTGIPVIAAGGFFDGRGLAAALSYGAAGVAMGTR
TH_1758|M.thermoresistible__bu PVPTTLLLPSVLDAVRDTGMPVVAAGGFFDGRGLAAALAYGAAGVAMGTR
Mflv_1487|M.gilvum_PYR-GCK PIATTLLLPSVLDAVADTGMPVIAAGGFFDGRGLAAALTYGAAGVAMGTR
Mvan_5269|M.vanbaalenii_PYR-1 PIATTLLLPSVLDAVADTGMPVIAAGGFFDGRGLAAALSYGAAGVAMGTR
MSMEG_6004|M.smegmatis_MC2_155 PIATTLLLPSVLDAVADTGMPVIAAGGFFDGRGLAAALSYGAAGVAMGTR
MAB_0603c|M.abscessus_ATCC_199 PVATTLLLPSVLDAVKDTGMPVVAAGGFFDGRGLAAALSYGAAGVAMGTR
*:.************ .**:**:********:****** ***********
Mb3583|M.bovis_AF2122/97 FLLTSDSTVPDAVKRRYLQAGLDGTVVTTRVDGMPHRVLRTELVEKLESG
Rv3553|M.tuberculosis_H37Rv FLLTSDSTVPDAVKRRYLQAGLDGTVVTTRVDGMPHRVLRTELVEKLESG
MMAR_5042|M.marinum_M FLLTSDSTVPEAVKRRYLQAALDGTVVTTRVDGMPHRVLRTGLVEKLEGG
MUL_4116|M.ulcerans_Agy99 FLLTSDSTVPDAVKRRYLQAALDGTVVTTRVDGMPHRVLRTGLVEKLEGG
MAV_0607|M.avium_104 FLLTSDSTVPDAVKKRYLEAALDGTVVTTRVDGMPHRVLRTGLVEKLESG
TH_1758|M.thermoresistible__bu FLLTKDSTVPDAVKRRYLEAGLDGTVVSTRVDGMPHRVLRTGLVEKLESG
Mflv_1487|M.gilvum_PYR-GCK FLLTKDSTVPDAVKQRYLDAALDGTVVSTKVDGMPHRVLRTGLVEKLESG
Mvan_5269|M.vanbaalenii_PYR-1 FLLTSDSTVPDAVKQRYLDAALDGTVVSTKVDGMPHRVLRTGLVEKLESG
MSMEG_6004|M.smegmatis_MC2_155 FLLTSDSTVPDAVKQRYLQAALDGTVVSTRVDGMPHRVLRTGLVEKLESG
MAB_0603c|M.abscessus_ATCC_199 FLLTSDSSVPDGVKQRYLASDLAGTVVSTRVDGMPHRVLRTELVEKLESG
****.**:**:.**:*** : * ****:*:*********** ******.*
Mb3583|M.bovis_AF2122/97 SRARGFAAALRNAGKFRRMSQMTWRSMIRDGLTMRHGKELTWSQVLMAAN
Rv3553|M.tuberculosis_H37Rv SRARGFAAALRNAGKFRRMSQMTWRSMIRDGLTMRHGKELTWSQVLMAAN
MMAR_5042|M.marinum_M SRARGFTAAIRNAAKFKQMSQMSWASMIKDGLAMRHGKELTWSQVVMAAN
MUL_4116|M.ulcerans_Agy99 SRARGFTAAIRNAAKFKQMSQMSWASMIKDGLAMRHGKELTWSQVLMAAN
MAV_0607|M.avium_104 SPVRGLTAAVRNAAKFKHMSKMTWRSMISDGLAMRHGKEMTWSQVVMAAN
TH_1758|M.thermoresistible__bu SPIRGFTAAVRNAQKFKKMTGMTWRSMIRDGLEMRRGKDLTWAQVIMAAN
Mflv_1487|M.gilvum_PYR-GCK SPVRGLSAAVLNAQKFKKMSGMTWKSMITDGLAMRHGKDLTWSQVVMAAN
Mvan_5269|M.vanbaalenii_PYR-1 SPIRGLSAAVLNAQKFKKMSGMTWKSMITDGLAMRHGKDLTWSQVVMAAN
MSMEG_6004|M.smegmatis_MC2_155 SRVRGFAAAVGNAQKFKKMSGMTWRSMIKDGLAMRHGKELTWSQVVMAAN
MAB_0603c|M.abscessus_ATCC_199 SRVRGFTAAVRNAAKFKKMTGMSWASMVKDGLAMRHGKELTWSQVLMAAN
* **::**: ** **::*: *:* **: *** **:**::**:**:****
Mb3583|M.bovis_AF2122/97 TPMLLKAGLVDGNTEAGVLASGQVAGILDDLPSCKELIESIVLDAITHLQ
Rv3553|M.tuberculosis_H37Rv TPMLLKAGLVDGNTEAGVLASGQVAGILDDLPSCKELIESIVLDAITHLQ
MMAR_5042|M.marinum_M TPMLLKAGLVDGNPEAGVLASGQVAGILDDLPSCAQLVESIVHDAIEHLR
MUL_4116|M.ulcerans_Agy99 TPMLLKAGLVDGNPEAGVLASGQVAGILDDLPSCAQLVESIVHDAIEHLR
MAV_0607|M.avium_104 TPMLLKAGLVEGNTEAGVLASGQVAGILEDLPSCAELIDSIVRDAIAHLR
TH_1758|M.thermoresistible__bu TPMLLKAGLVEGNTEAGVLASGQVAGILDDLPSCAELIEQIVADAVRHLK
Mflv_1487|M.gilvum_PYR-GCK TPMLLKAGLVEGNTEAGVLASGQVAGIIDDLPSCAELVPAIVAEAVEHLQ
Mvan_5269|M.vanbaalenii_PYR-1 TPMLLKAGLVEGNTDAGVLASGQVAGIIDDLPSCAELVPAIVAEAVEHLQ
MSMEG_6004|M.smegmatis_MC2_155 TPMLLKAGLVEGNTDAGVLASGQVTGIIDDLPSCAELVPAIVADAVEHLN
MAB_0603c|M.abscessus_ATCC_199 TPMLLKAGLVEGNTNAGVLASGQVAGILDDLPSCHELIDAIVTDAVAHLK
**********:**.:*********:**::***** :*: ** :*: **.
Mb3583|M.bovis_AF2122/97 TASALVE--
Rv3553|M.tuberculosis_H37Rv TASALVE--
MMAR_5042|M.marinum_M AAAALVE--
MUL_4116|M.ulcerans_Agy99 AAAALVE--
MAV_0607|M.avium_104 SASGLIEE-
TH_1758|M.thermoresistible__bu GAVALVEE-
Mflv_1487|M.gilvum_PYR-GCK KASQYIR--
Mvan_5269|M.vanbaalenii_PYR-1 KASQHIR--
MSMEG_6004|M.smegmatis_MC2_155 KASSFIQEK
MAB_0603c|M.abscessus_ATCC_199 AANATIL--
* :