For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HRIPRPLRQHWKLLGAFVVFVAVVALVFGDVRIRPYITGDPTVIASEITENIAGTVDLFDNTVPHSLSVE LTDAEYRDMVEQFKADGDKKWVTADITIDGTTINDVAVRLKGNSTLMSLRGVHFPPPPGEGGPPGPPGPP GRPGPPGPPGAPGPGGPPGGFGGPGGLGGPGGPMGTVSEDDPLSLPLLISFNENADGRGYQGLTELSVRP GAPVLNEALALSLTAMTGQPTQRYSYITYSVNGATTTRLLLEHPDENYASTVFDSDGYLYKARAKSRLEY AGPDQSAYADQFKQINTVDTGNLQPLINFLKWLDSADDEEFARSLGDWIDVESFARYVATQNLLINGDDM AGPGQNYYLWYDLGTKKFTVVSWDLNLATMMGDVKLGPRDPMEFKPPSGFQLPTGMGPPPGKELGGNILK ERFLASPAFTEVYDKAYWELYDQIYADGQALALLDSIAETVPVTDGLSADEIEAQKDTLRKFLTERASAL AKVRES*
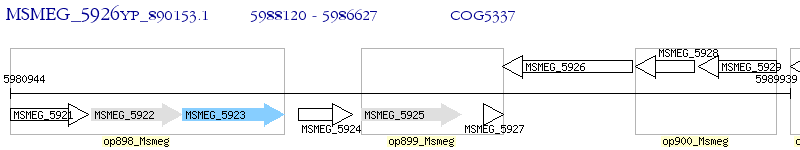
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5926 | - | - | 100% (497) | putative secreted protein |
| M. smegmatis MC2 155 | MSMEG_3545 | - | 2e-11 | 58.62% (58) | hypothetical protein MSMEG_3545 |
| M. smegmatis MC2 155 | MSMEG_5268 | - | 3e-11 | 57.14% (56) | hypothetical protein MSMEG_5268 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1485c | PE_PGRS27 | 2e-08 | 50.00% (66) | PE-PGRS family protein |
| M. gilvum PYR-GCK | Mflv_2044 | - | 4e-09 | 55.17% (58) | integral membrane protein-like protein |
| M. tuberculosis H37Rv | Rv1450c | PE_PGRS27 | 1e-07 | 55.32% (47) | PE-PGRS family protein |
| M. leprae Br4923 | MLBr_02395 | Ag36 | 2e-05 | 51.22% (41) | proline rich antigenic protein |
| M. abscessus ATCC 19977 | MAB_2532 | - | 7e-09 | 59.26% (54) | hypothetical protein MAB_2532 |
| M. marinum M | MMAR_3811 | - | 2e-11 | 60.71% (56) | hypothetical protein MMAR_3811 |
| M. avium 104 | MAV_3436 | - | 3e-11 | 55.00% (60) | hypothetical protein MAV_3436 |
| M. thermoresistible (build 8) | TH_0385 | - | 1e-171 | 63.95% (466) | putative secreted protein |
| M. ulcerans Agy99 | MUL_1750 | - | 6e-10 | 48.44% (64) | proline and glycine rich transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_5408 | - | 2e-09 | 44.30% (79) | hypothetical protein Mvan_5408 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 MSLVIVAPETVAAAALDVARIGSSIGVANSAAAGSTTSVLAAGADEVSAA
Rv1450c|M.tuberculosis_H37Rv MSLVIVAPETVAAAALDVARIGSSIGAANAAAAGSTTSVLAAGADEVSAA
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 IATLFGSHAREYQAISTQVAAFHDRFAQTLSAAVGSYVSAEATNAAPLAT
Rv1450c|M.tuberculosis_H37Rv IATLFGSHAREYQAISTQVAAFHDRFAQTLSAAVGSYVSAEATNAAPLAT
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 LEHNVLNALNAPTQALLGRPLIGDGAAGAPGTGQAGGAGGILWGNGGAGG
Rv1450c|M.tuberculosis_H37Rv LEHNVLNALNAPTQALLGRPLIGDGAAGAPGTGQAGGAGGILWGNGGAGG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 SGAPGQVGGAGGAAGLFGTGGAGGAGGAGAAGGAGGSGGWLLGNGGVGGA
Rv1450c|M.tuberculosis_H37Rv SGAPGQVGGAGGAAGLFGTGGAGGAGGAGAAGGAGGSGGWLLGNGGVGGA
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GGQSLLGGATGGAGGNAGLFGVGGTGGPGGPGGPGGPGGPGGPGGVGGTG
Rv1450c|M.tuberculosis_H37Rv GGQSLLGGATGGAGGNAGLFGVGGTGGPGGPGG---------PGGVGGTG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GAGGLGGTLYGAGGHGGAGGPGPIGGVGGHGGVGGAAGLLGVGGHGGAGG
Rv1450c|M.tuberculosis_H37Rv GAGGLGGTLYGAGGHGGAGGPGPIGGVGGHGGVGGAAGLLGVGGHGGAGG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 HGAAGIAGAAGKGGIFPDGSNGGAGGDAGDGGTGGRGGWLAGAGGAGGDG
Rv1450c|M.tuberculosis_H37Rv HGAEGVAGAAG-EDLSPHGTSGGVGGDAGDGGTGGRGGWLAGAGGAGGAG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GIGGTGGAGGAGFSRALIVAGDNGGDGGNGGMGGAGGAGGPGGAGGLISL
Rv1450c|M.tuberculosis_H37Rv GVGGTGGAGGAGFSRALIVAGDNGGD------------------------
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu -----------------------------------------------MDR
Mb1485c|M.bovis_AF2122/97 LGGQGAGGDGGDGGAGGVGGDRGAGANGNQAVNAGAGGAGGHGGDPGAGG
Rv1450c|M.tuberculosis_H37Rv ---------------------------------------------PGAGG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 -------MHRIPRPLRQHWKLLGAFVVFVAVVALVFGDVRIRPYITGDPT
TH_0385|M.thermoresistible__bu PGLPRRLVRRLPKPLRQHWKPLVATVAVLAFVAVWFGDVRIRPYVTGAVH
Mb1485c|M.bovis_AF2122/97 AGGTGGAGSTIGAHGAAGASPTSGGNGGAGGNGAHFSSGGKAGGNGGAGG
Rv1450c|M.tuberculosis_H37Rv AGGTGGAGSTIGAHGAAGASPTSGGNGGAGGNGAHFSSGGKAGGNGGAGG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 ----------------MSQPPEYPGTPPEP------PPGYGPPPGYG---
MUL_1750|M.ulcerans_Agy99 ----------------MSQPPENPGNQADPQDGDQSTPGYPPPPNYGTPP
Mflv_2044|M.gilvum_PYR-GCK ----------------MTENPPPGGYPPPP------QGGYPPPPPSEGGY
MLBr_02395|M.leprae_Br4923 ---------------MTDQPPPSGSNPTPAPPPPGSSGGYEPSFAPSELG
MSMEG_5926|M.smegmatis_MC2_155 VIASEITENIAGTVDLFDNTVPHSLSVELTDAEYRDMVEQFKADGDKKWV
TH_0385|M.thermoresistible__bu VAAPRITHDIAGTVDLFDPTVAHTVSVEMSDAQYRDMIAQYAADGDKKWM
Mb1485c|M.bovis_AF2122/97 AGGLVGNGGAGGAGGNGAPGAPPSGGDPNGGGGGAGGAGGKGGDGGAQAG
Rv1450c|M.tuberculosis_H37Rv AGGLVGNGGAGGAGGNGAPGAPPSGGDPNGGGGGAGGAGGKGGDGGAQAG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --PPPGYG-------------PPPGYGTPPPPPP-GYG------PPPGYG
MUL_1750|M.ulcerans_Agy99 PPPPPGYGAPPAGPPGYGAPPPPPGYGTPPPPPPPGYGQAPGGYAPPGYN
Mflv_2044|M.gilvum_PYR-GCK PPPPPEGGYPPPPPAG-GYQQPPPGGAYPPPPGPGGYP------PPPGQG
MLBr_02395|M.leprae_Br4923 SAYPPPTAPPVG---GSYPPPPPPGGSYPPPPPPGGSYPP----PPPSTG
MSMEG_5926|M.smegmatis_MC2_155 TADITIDGTTINDVAVRLKGNSTLMSLRGVHFPPPPGEGGPPGPPGPPGR
TH_0385|M.thermoresistible__bu TARVTIDGTVIDDVAVRLKGNSTLMGLRGDGF-------------GPPGW
Mb1485c|M.bovis_AF2122/97 DGGAGGAGGKGGNGGNGATGATGLNGLGAGADGTDGGKGGNGGAGGGGGA
Rv1450c|M.tuberculosis_H37Rv DGGAGGAGGKGGNGGNGATGATGLNGLGAGADGTDGGKGGNGGAGGGGGA
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 ---APPPGYGPPP-GYG---PPPGF-------------------------
MUL_1750|M.ulcerans_Agy99 PPPPPPPGYGPPPAGYS---TQPGF-------------------------
Mflv_2044|M.gilvum_PYR-GCK GYPPPPGGYGMPPAGFGGGYPPPGQ-------------------------
MLBr_02395|M.leprae_Br4923 AYAPPPPG------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 PGPPGPPGAPGPGGPPGGFGGPGGLGGPGGPMGTVSEDDP----------
TH_0385|M.thermoresistible__bu SGPP-PSKAP---------------------FSAVSADDP----------
Mb1485c|M.bovis_AF2122/97 GGQGGKALAATHQDGSMGAGGAGGNGGAGGMGGDGGNGAKGTFDNGGDGV
Rv1450c|M.tuberculosis_H37Rv GGQGGKALAATHQDGSMGAGGAGGNGGAGGMGGDGGNGAKGTFDNGGDGV
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 ------------LSLPLLISFNENADGRGYQGLTELSVRPGAPVLNEALA
TH_0385|M.thermoresistible__bu ------------TSLPLLLSFDEHAAGRAYQGMTELAVRPGAPVLNEALA
Mb1485c|M.bovis_AF2122/97 GGNGGNGGSRGIGGAGGIGGAGSTAGADGARGATPTSGGNGGTGGNGANA
Rv1450c|M.tuberculosis_H37Rv GGNGGNGGSRGIGGAGGIGGAGSTAGADGARGATPTSGGNGGTGGNGANA
MMAR_3811|M.marinum_M ---------------------------------MKTYPRLAATG----LA
MAB_2532|M.abscessus_ATCC_1997 ---------------------------------MHTKARTTVGVAVGSMA
Mvan_5408|M.vanbaalenii_PYR-1 -------------MTETPESRTEPTAVATEQGRGELSPGSDRPNRLTQSA
MAV_3436|M.avium_104 -------------GGPPK---PAFSVGEAFGWAWNAFTKNPVALIVPTLV
MUL_1750|M.ulcerans_Agy99 -------------GGPAK---PPFSVGEAISWAWNRFTQNAMALVVPIVI
Mflv_2044|M.gilvum_PYR-GCK -------------GYPPVGGPPSLDIGAAFSWSFNKFSKNAVPLIVPTLV
MLBr_02395|M.leprae_Br4923 -----------------------PAIRSLPKEAYTFWVTRVLAYVIDNIP
MSMEG_5926|M.smegmatis_MC2_155 LSLT--------------------------AMTGQPTQRYSYITYSVN--
TH_0385|M.thermoresistible__bu LTMT--------------------------ALTGQPTQRYAYVTYSIN--
Mb1485c|M.bovis_AF2122/97 TVAGGAGGAGGKGGNGGLVGNGGAGGKGGDGMAGVAGSSPTTAGESGTSG
Rv1450c|M.tuberculosis_H37Rv TVAGGAGGAGGKGGNGGLVGNGGAGGKGGDGMAGVAGSSPTTAGESGTSG
MMAR_3811|M.marinum_M MVLG-------------------------LGLAGVG-----VASEA----
MAB_2532|M.abscessus_ATCC_1997 LIAS---------------------------LIGCGTDAPPRSVGT----
Mvan_5408|M.vanbaalenii_PYR-1 AWVG--------------------------IVAGVVFIVAVIFFSG----
MAV_3436|M.avium_104 Y----------------------------LVVLGGAST--LIGLSQDV--
MUL_1750|M.ulcerans_Agy99 YGLI-------------------------LSAVGGVMVGLFFAFSDRT--
Mflv_2044|M.gilvum_PYR-GCK YALV-------------------------IGVLGAVIFG-LASLFPAD--
MLBr_02395|M.leprae_Br4923 ATVLLG--------------------------IGMLIQTLTKQEACVT--
*
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------GATTTR
TH_0385|M.thermoresistible__bu --------------------------------------------GATTTR
Mb1485c|M.bovis_AF2122/97 QNGGAGGAGGAGGRGGDFGGDGGTGGAGGNGADGGAGGNGANGANATTPG
Rv1450c|M.tuberculosis_H37Rv QNGGAGGAGGAGGRGGDFGGDGGTG---------GAGGNGANGANATTPG
MMAR_3811|M.marinum_M ---------------------------------------------AAQPG
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------TTQTPA
Mvan_5408|M.vanbaalenii_PYR-1 ------------------------------------------FFVGKQYG
MAV_3436|M.avium_104 ---------------------------------------------GTS-G
MUL_1750|M.ulcerans_Agy99 ---------------------------------------------STTYT
Mflv_2044|M.gilvum_PYR-GCK ---------------------------------------------YTSYS
MLBr_02395|M.leprae_Br4923 --------------------------------------------DITQYN
MSMEG_5926|M.smegmatis_MC2_155 LLLEHPDENYASTVFDSDGYLYKAR-------------------------
TH_0385|M.thermoresistible__bu LVLEHPDEGYPARVFGTDGYLYKAR-------------------------
Mb1485c|M.bovis_AF2122/97 AKGGDGGHGGPGAQGGNGGQGGPGGLAGNLFGQNGIQGVGGSGGKGGAGG
Rv1450c|M.tuberculosis_H37Rv AKGGDGGHGGPGAQGGNGGQGGPGGLAGNLFGQNGIQGVGGSGGKGGAGG
MMAR_3811|M.marinum_M AP----------TQWCPGDFWDPG------WGQN----------------
MAB_2532|M.abscessus_ATCC_1997 PP--------------PGGS------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 GG-----------YRDGRGMSYPGG-------------------------
MAV_3436|M.avium_104 GGSGDDYFTFTANLNGGGMALLVLG-------------------------
MUL_1750|M.ulcerans_Agy99 DAYGNTSETVNMTMSPLASIVMFIG-------------------------
Mflv_2044|M.gilvum_PYR-GCK GADG---AGMSLDMGPAATIILFLG-------------------------
MLBr_02395|M.leprae_Br4923 VNQYCATQPTGIGMLAFWFAWLMAT-------------------------
MSMEG_5926|M.smegmatis_MC2_155 ---------------AKSRLEYAGPDQSAYADQFKQIN------------
TH_0385|M.thermoresistible__bu ---------------AGSRLEYRGPSQAAYAHEFKQVN------------
Mb1485c|M.bovis_AF2122/97 LAGDGGNGANGNFAFGDGNGGHGGNGGNPGAGGQGGSGGAGSTPGAKGAH
Rv1450c|M.tuberculosis_H37Rv LAGDGGNGANGNFAFGDGNGGHGGNGGNPGAGGQGGSGGAGSTPGAKGAH
MMAR_3811|M.marinum_M ------------WDMGHCHDNWRGPGPNP---------------------
MAB_2532|M.abscessus_ATCC_1997 ---------------GSVPGGPTGGGGPEG--------------------
Mvan_5408|M.vanbaalenii_PYR-1 ---------------MMGPGGMMGPGGMMGP-------------------
MAV_3436|M.avium_104 ------------YLVAYLVGAFAQSAYLSG--------------------
MUL_1750|M.ulcerans_Agy99 ------------YLAVFAVVLFMHAGITTG--------------------
Mflv_2044|M.gilvum_PYR-GCK ------------LIMLFVVGGAISAAYMAG--------------------
MLBr_02395|M.leprae_Br4923 ---------------AYLVWNYGYRQGATG--------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GFTPTSGGDGGDGGNGGHSQVVGGNGGDGGNGGNGGSAGTGGNGGRGGDG
Rv1450c|M.tuberculosis_H37Rv GFTPTSGGDGGDGGNGGNSQVVGGNGGDGGNGGNGGSAGTGGNGGRGGDG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AFGGMSANATNPGENGPNGNPGGNGGAGGAGGAGLNGGNGGAGGNGGLGG
Rv1450c|M.tuberculosis_H37Rv AFGGMSANATNPGENGPNGNPGGNGGAGGAGGAGLNGGNGGAGGNGGLGG
MMAR_3811|M.marinum_M ----------QPGHAGP---------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 -----------TVDTGNLQPLINFLKWLDSADDEEFARSLGDWIDVESFA
TH_0385|M.thermoresistible__bu -----------AAETGNLQPLIDFLHWLDTADDDAFAAGLSRWVDVESFA
Mb1485c|M.bovis_AF2122/97 FGGNGAAGANGVAVGAPGQPGGAGGHGGAGGNGGAGGNGGQGVVSDGAGG
Rv1450c|M.tuberculosis_H37Rv FGGNGAAGANGVAVGAPGQPGGAGGHGGAGGNGGAGGNGGQGVVSDGAGG
MMAR_3811|M.marinum_M --------------GGPGEP--APGQQGPGGPGGPGGPG------EGPGG
MAB_2532|M.abscessus_ATCC_1997 --------------GAGSIPGGPTGGGGPGGGGGEIPGG-----PTGSGG
Mvan_5408|M.vanbaalenii_PYR-1 --------------GGMMGPGGPGGMMGPGGPGGMMGPG-----SWGPGG
MAV_3436|M.avium_104 -----------CLDLADGRPVTIGSFFKPRNFGMVFLAALLVGILTSIAS
MUL_1750|M.ulcerans_Agy99 -----------CLDIADGKPVTIGSFFKPRNLGMVILTGLLVIVLTAIGS
Mflv_2044|M.gilvum_PYR-GCK -----------VLDIANGQQVEFGSFFKPRNIGAVVIASLIVGIATSIGY
MLBr_02395|M.leprae_Br4923 -------------SSIGKTVMKFKVISEATGQPIGFGMSVVRQLAHFVDA
.
MSMEG_5926|M.smegmatis_MC2_155 RYVATQNLLIN------------------------------GDDMAGPGQ
TH_0385|M.thermoresistible__bu RYVATQNLLVN------------------------------ADDMAGPGQ
Mb1485c|M.bovis_AF2122/97 AGGAGGDGGAPGDGANGGNGQGAGAFAGGGGGRGGDGGNAGNAGAGGPGG
Rv1450c|M.tuberculosis_H37Rv AGGAGGDGGAPGDGANGGNGQGAGAFAGGGGGRGGDGGNAGNAGAGGPGG
MMAR_3811|M.marinum_M PGGPG-------------------------------------EGPGGPGG
MAB_2532|M.abscessus_ATCC_1997 PGGGSG----------------------------------CIPNVGCVNV
Mvan_5408|M.vanbaalenii_PYR-1 MMGPGSWG----------------------------------PGMMGPGQ
MAV_3436|M.avium_104 ALCFLPGLILG-----------------------------LFAQFTIAYA
MUL_1750|M.ulcerans_Agy99 VLCIVPGLIFG-----------------------------FLAQFAIIAA
Mflv_2044|M.gilvum_PYR-GCK VLCIVPGLIVS-----------------------------IFALFTTVFI
MLBr_02395|M.leprae_Br4923 VICCIG--------------------------------------FLFPLW
MSMEG_5926|M.smegmatis_MC2_155 NYYLWYDLGTKKFTVVSWDLNLATMMGDVKLGPRDPMEFKPPSGFQLPTG
TH_0385|M.thermoresistible__bu NYYLWYDLATRRFTVLSWDLNLAMVMGDPDLGPRDPLKMKPPPGMPPPPQ
Mb1485c|M.bovis_AF2122/97 TGSTAGKAGPAGSILHDGGNGGHGGHGAASGGNGGPGGHGGNGGNGGTGA
Rv1450c|M.tuberculosis_H37Rv TGSTAGKAGPAGSILHDGGNGGHGGHGAASGGNGGPGGHGGNGGNGGTGA
MMAR_3811|M.marinum_M P--------------H----------------------------------
MAB_2532|M.abscessus_ATCC_1997 P-------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 QPSPTTTPAVPTTPQR----------------------------------
MAV_3436|M.avium_104 IDRSESAVKALSSSFSTVTANLANALLVWLAEFALVVVGALACGVGLLLA
MUL_1750|M.ulcerans_Agy99 VDRSLSPIDSIKSSCATVRAELGNTALSWLVQYAVVLAGELLCFVGMLVG
Mflv_2044|M.gilvum_PYR-GCK VDRNLSAIDGIKASIAVAKANFLQVFLTWLIFNVLISVGSFVCYIGLIVT
MLBr_02395|M.leprae_Br4923 DSKRQTLADKIMTTVCLPI-------------------------------
MSMEG_5926|M.smegmatis_MC2_155 MGPPPGKE--LGGNILKERFLASPAFTEVYDKAYWELYDQIYADGQALAL
TH_0385|M.thermoresistible__bu DWPPSGGPPWREGNLLKERFLAAPAFDELYQTVYWELYDQLYADGRAVDL
Mb1485c|M.bovis_AF2122/97 NGGNGGIGGTGGAGSTGAKGVLGTNEGDGGDGGRGGNGGRGGNGGQGLTG
Rv1450c|M.tuberculosis_H37Rv NGGNGGIGGTGGAGSTGAKGVLGTNEGDGGDGGRGGNGGRGGNGGQGLTG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 APVAALVGIYAYRKLSGGQVAPPPGYQPGPPPGYQPGPPPGAPPA-----
MUL_1750|M.ulcerans_Agy99 VPVASLIQTYTWRKLSGGQVVELS--QPGPPAGLPPGPPPGAPPGPQFG-
Mflv_2044|M.gilvum_PYR-GCK VPLAVLYMVYAYRTLTGGYVAPAT---P----------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 LDSIAETVPVTDGLSADEIEAQKDTLRKFLTERASALAKVRES-------
TH_0385|M.thermoresistible__bu LDQVVAAVPVXX-------------------DPPSAGAHRR---------
Mb1485c|M.bovis_AF2122/97 AGGNGGTGGTPGNGGNGGNGASGDLVTSPGDGGGGGRGGDAGRGGDAGLG
Rv1450c|M.tuberculosis_H37Rv AGGNGGTGGTPGNGGNGGNGASGDLVTSPGDGGGGGRGGDAGRGGDAGLG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GSSGPGGTPGDWGTGGTGGTGGTGGQGANGGLTGGRGGTGGNGG------
Rv1450c|M.tuberculosis_H37Rv GSSGPGGTPGDWGTGGTGGTGGTGGQGANGGLTGGRGGTGGNGGNGNTGG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 ---AGGTGGTGHNGSQPGMGGNGGAGGFGGNGFAGVGGRGGMGGSGGTGG
Rv1450c|M.tuberculosis_H37Rv TGGAGGTGGTGHNGSQPGMGGNGGAGGFGGNGFAGVGGRGGMGGSGGTGG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 --------------------------------------------------
TH_0385|M.thermoresistible__bu --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 TGDAGPFGTGTGGTGGHGGQGGGGGFSILLGLGGLGGLGSPGSIATGTAG
Rv1450c|M.tuberculosis_H37Rv TGDAGPFGTGTGGTGGHGGQGGGGGFSILLGLGGLGGLGSPGSIATGTAG
MMAR_3811|M.marinum_M --------------------------------------------------
MAB_2532|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_5408|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3436|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2044|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02395|M.leprae_Br4923 --------------------------------------------------
MSMEG_5926|M.smegmatis_MC2_155 -----------------
TH_0385|M.thermoresistible__bu -----------------
Mb1485c|M.bovis_AF2122/97 GAGGGGGFGGLGGGEFV
Rv1450c|M.tuberculosis_H37Rv GAGGGGGFGGLGGGEFV
MMAR_3811|M.marinum_M -----------------
MAB_2532|M.abscessus_ATCC_1997 -----------------
Mvan_5408|M.vanbaalenii_PYR-1 -----------------
MAV_3436|M.avium_104 -----------------
MUL_1750|M.ulcerans_Agy99 -----------------
Mflv_2044|M.gilvum_PYR-GCK -----------------
MLBr_02395|M.leprae_Br4923 -----------------