For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDQPPGNNPPPPDGGYPPPPPPDGGYPPPPPPDGGYPPAQPGGFGPPPQGGYPPPPPPGGYPPPPQGGFP PPPPGGYPPPPPPQGGSYPPPPPPGAAGYPPPGYPGGPGAGYPPAGGFGQPGGYAPVGAPGFGGAYSVGD AFSWAWNKFSKHAVEMIVPALVFGLVYAILQGIVNGISGAFTSTSSADGFELSMATGGGVVSIIGAIITL IVTAVIQAAYISGVLEIANGQPVTIGSFFKPRNVGDVIIATVIVGVINFVVAAILLFPGFFVPGYLFVGV PVLLIASAIIAVLFLFTTVAVLDRNLSGVDAVKTSFELSKANFGTVFITAVVIFLLLLAGAIACGIGLLV AYPLVALIEVYAFRRLTGAPIAPPTQ*
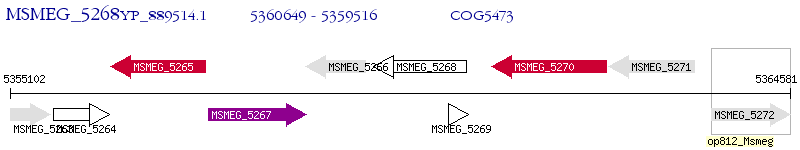
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5268 | - | - | 100% (377) | hypothetical protein MSMEG_5268 |
| M. smegmatis MC2 155 | MSMEG_0035 | - | 8e-21 | 40.83% (169) | FHA domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5895 | - | 1e-14 | 35.17% (145) | virulence factor mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2590 | - | 1e-50 | 32.14% (364) | hypothetical protein Mb2590 |
| M. gilvum PYR-GCK | Mflv_2044 | - | 1e-93 | 48.40% (376) | integral membrane protein-like protein |
| M. tuberculosis H37Rv | Rv2560 | - | 1e-50 | 32.14% (364) | hypothetical protein Rv2560 |
| M. leprae Br4923 | MLBr_02395 | Ag36 | 3e-22 | 47.32% (112) | proline rich antigenic protein |
| M. abscessus ATCC 19977 | MAB_0651c | - | 3e-43 | 34.75% (377) | hypothetical protein MAB_0651c |
| M. marinum M | MMAR_2166 | - | 5e-70 | 38.64% (396) | proline and glycine rich transmembrane protein |
| M. avium 104 | MAV_3436 | - | 8e-66 | 40.53% (338) | hypothetical protein MAV_3436 |
| M. thermoresistible (build 8) | TH_4441 | - | 1e-52 | 39.77% (259) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1750 | - | 2e-66 | 37.08% (391) | proline and glycine rich transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_4673 | - | 2e-92 | 50.40% (375) | hypothetical protein Mvan_4673 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2590|M.bovis_AF2122/97 -MSQ------------PPEHPGNPADPQGG--NQGAG-------------
Rv2560|M.tuberculosis_H37Rv -MSQ------------PPEHPGNPADPQGG--NQGAG-------------
MMAR_2166|M.marinum_M -MSQ------------PPENPGNQADPQGG--DQSTP-------------
MUL_1750|M.ulcerans_Agy99 -MSQ------------PPENPGNQADPQDG--DQSTP-------------
MAV_3436|M.avium_104 -MSQ------------PPEYPGTPPEPPPG--YGPPP-------------
MAB_0651c|M.abscessus_ATCC_199 -MTY------------PPQEPPTSGDPSAYDPNQPPP-------------
MSMEG_5268|M.smegmatis_MC2_155 -MTDQPPGN------NPPPPDGGYPPPPPPDGGYPPPPPPDGGYPPAQPG
Mvan_4673|M.vanbaalenii_PYR-1 -MTE-----------NPPP--GGYPPPPP--GGYPPPPPP---------G
Mflv_2044|M.gilvum_PYR-GCK -MTE-----------NPPP--GGYPPPPQ--GGYPPPPPSE--------G
TH_4441|M.thermoresistible__bu MLSAVFIGRGVDAVRSPKPAADAARPTIEGLTKLPEPVGPR--------V
MLBr_02395|M.leprae_Br4923 -MTD------------QPPPSGSNPTPAPPPPGSSG--------------
:: .
Mb2590|M.bovis_AF2122/97 SYPPP----------------------GYGAPPPPP--------------
Rv2560|M.tuberculosis_H37Rv SYPPP----------------------GYGAPPPPP--------------
MMAR_2166|M.marinum_M GYPPPPNYGTPPPPPPPGYGAPPAGPPGYGAPPPPPGYGTPPPPPGYGTP
MUL_1750|M.ulcerans_Agy99 GYPPPPNYGTPPPPPPPGYGAPPAGPPGYGAPPPPP---------GYGTP
MAV_3436|M.avium_104 GYGPPPGYGPPP---------------GYGTPPPPP--------------
MAB_0651c|M.abscessus_ATCC_199 GYPNQP---------------------PPGYQPPPP--------------
MSMEG_5268|M.smegmatis_MC2_155 GFGPPPQ-GGYPPPPPPGGYP----PPPQGGFPPPP--------------
Mvan_4673|M.vanbaalenii_PYR-1 GYQPPP------PPPSGGGYP----PPPSGGFPPPQ--------------
Mflv_2044|M.gilvum_PYR-GCK GYPPPPPEGGYPPPPPAGGYQQ---PPPGGAYPPPPG-------------
TH_4441|M.thermoresistible__bu PTDAETVARINGMVQIAGGAL-----LATGKLPRVAS-------------
MLBr_02395|M.leprae_Br4923 GYEPSFAP------------------SELGSAYPPP--------------
*
Mb2590|M.bovis_AF2122/97 ------GYGPPPGTYLPPG---YN-APPPP----PGYGPPPG-------P
Rv2560|M.tuberculosis_H37Rv ------GYGPPPGTYLPPG---YN-APPPP----PGYGPPPG-------P
MMAR_2166|M.marinum_M PPPPPPGYGQAPGGYAPPG---YNPPPPPP----PGYGPPPAG-----YP
MUL_1750|M.ulcerans_Agy99 PPPPPPGYGQAPGGYAPPG---YNPPPPPP----PGYGPPPAG-----YS
MAV_3436|M.avium_104 -----PGYGP------PPG---YG---APP----PGYGPPPG------YG
MAB_0651c|M.abscessus_ATCC_199 ------GYQPGYQAGPPPGGPQYQGAQFPP----PGY--PPAG-----YG
MSMEG_5268|M.smegmatis_MC2_155 PGGYPPPPPPQGGSYPPPPPPGAAGYPPPGYPGGPGAGYPPAGGFGQPGG
Mvan_4673|M.vanbaalenii_PYR-1 EGGYPPPQGP--GGYPPP-----AGYGPP------QGGFPPP-----AQG
Mflv_2044|M.gilvum_PYR-GCK PGGYPPPPGQ--GGYPPPP----GGYGMP--PAGFGGGYPPP-----GQG
TH_4441|M.thermoresistible__bu AVLALSVVPGSLGGHMFWTESDPQRKAXX-----XTGGYPPA-------G
MLBr_02395|M.leprae_Br4923 ------TAPPVGGSYPPPPPPGGSYPPPPP----PGGSYPPP--------
**
Mb2590|M.bovis_AF2122/97 PPPGYPTHLQSSGFSVGDAISWSWNRFTQNAVTLVVPVLAYAVALAAVIG
Rv2560|M.tuberculosis_H37Rv PPPGYPTHLQSSGFSVGDAISWSWNRFTQNAVTLVVPVLAYAVALAAVIG
MMAR_2166|M.marinum_M AQPGFGGPAKPP-FSVGEAISWAWNRFTQNAMALVVPIVIYGLILSAVGG
MUL_1750|M.ulcerans_Agy99 TQPGFGGPAKPP-FSVGEAISWAWNRFTQNAMALVVPIVIYGLILSAVGG
MAV_3436|M.avium_104 PPPGFGGPPKPA-FSVGEAFGWAWNAFTKNPVALIVPTLVYLVVLGGAS-
MAB_0651c|M.abscessus_ATCC_199 YPQQRPWPPQGP-FSAGESWSWSWAQVSKRFGTFIPPYLVWFLAIGLPVG
MSMEG_5268|M.smegmatis_MC2_155 YAPV-GAPGFGGAYSVGDAFSWAWNKFSKHAVEMIVPALVFGLVYAILQG
Mvan_4673|M.vanbaalenii_PYR-1 YPPV-GGP---PAYSVGDAFNWAWNKFSKNAMPLIVATLVFGIVVIALQA
Mflv_2044|M.gilvum_PYR-GCK YPPV-GGP---PSLDIGAAFSWSFNKFSKNAVPLIVPTLVYALVIGVLGA
TH_4441|M.thermoresistible__bu FPPP-GGFG--ASYRFGDAFNWAWSAFTRHAAPLMLAALAYVAIIFALSF
MLBr_02395|M.leprae_Br4923 PPSTGAYAPPPPGPAIRSLPKEAYTFWVTRVLAYVIDNIPATVLLGIGML
:: . : :
Mb2590|M.bovis_AF2122/97 ATAGLVVALSDRATTAYTNTSGVSSESVDIT---MTPAAGIVMFLGYIAL
Rv2560|M.tuberculosis_H37Rv ATAGLVVALSDRATTAYTNTSGVSSESVDIT---MTPAAGIVMFLGYIAL
MMAR_2166|M.marinum_M VMVGLFFAFSDRTSTTYTDAYGNTSETVNMT---MSPLASIVMFIGYLAV
MUL_1750|M.ulcerans_Agy99 VMVGLFFAFSDRTSTTYTDAYGNTSETVNMT---MSPLASIVMFIGYLAV
MAV_3436|M.avium_104 ----TLIGLSQDVGTS-GGGSGDDYFTFTAN---LNGGGMALLVLGYLVA
MAB_0651c|M.abscessus_ATCC_199 IVYAILMASLPQTSTSGYGGNSRSSYSYSYEGPELSGGAIAIMILLYAVV
MSMEG_5268|M.smegmatis_MC2_155 IVNGISGAFTS------TSSADGFELSMATG-------GGVVSIIGAIIT
Mvan_4673|M.vanbaalenii_PYR-1 IIN-IVQALVSPGDTSYIADDSGFSFSYATTG----VAGTIVAIVGWFLS
Mflv_2044|M.gilvum_PYR-GCK VIFGLASLFPADYTSYSGADGAGMSLDMGPA-------ATIILFLGLIML
TH_4441|M.thermoresistible__bu LFSLVLTAFTPGG--ATTVDPDSFFDMFGVG-------GTLTMVVGVILL
MLBr_02395|M.leprae_Br4923 IQTLTKQEACVTDITQYNVNQYCATQPTGIG----------------MLA
Mb2590|M.bovis_AF2122/97 FALVLYMHAGILTGCLDIADGKPVTIATFFRPRNLGLVLVTGLLIVALTF
Rv2560|M.tuberculosis_H37Rv FALVLYMHAGILTGCLDIADGKPVTIATFFRPRNLGLVLVTGLLIVAVTF
MMAR_2166|M.marinum_M FAVVLFMHAGITTGCLDIADGKPVTIGSFFKPRNLGMVILTGLLVIVLTA
MUL_1750|M.ulcerans_Agy99 FAVVLFMHAGITTGCLDIADGKPVTIGSFFKPRNLGMVILTGLLVIVLTA
MAV_3436|M.avium_104 YLVGAFAQSAYLSGCLDLADGRPVTIGSFFKPRNFGMVFLAALLVGILTS
MAB_0651c|M.abscessus_ATCC_199 FAVSLYVGACLISANLDVADGKPVSFGTFFRARGFGLYVGAALLVGVGVL
MSMEG_5268|M.smegmatis_MC2_155 LIVTAVIQAAYISGVLEIANGQPVTIGSFFKPRNVGDVIIATVIVGVINF
Mvan_4673|M.vanbaalenii_PYR-1 LIVTAAIQSAFLGGIFDIANGQQVAVGSFFRPRNVGNVIIAGLIVGVITT
Mflv_2044|M.gilvum_PYR-GCK FVVGGAISAAYMAGVLDIANGQQVEFGSFFKPRNIGAVVIASLIVGIATS
TH_4441|M.thermoresistible__bu LFVSGVITSAFLNGLLAIADGQQIELASFFRPRNVGSVVIATVIVGVLSY
MLBr_02395|M.leprae_Br4923 FWFAWLMATAYLVWNYGYRQG--------ATGSSIGKTVMK------FKV
. : :* ..* .
Mb2590|M.bovis_AF2122/97 IGGLLCVIPG------------------LIFGFVAQFAVAFAVDRSTSPI
Rv2560|M.tuberculosis_H37Rv IGGLLCVIPG------------------LIFGFVAQFAVAFAVDRSTSPI
MMAR_2166|M.marinum_M IGSVLCIVPG------------------LIFGFLAQFAIIAAVDRSLSPI
MUL_1750|M.ulcerans_Agy99 IGSVLCIVPG------------------LIFGFLAQFAIIAAVDRSLSPI
MAV_3436|M.avium_104 IASALCFLPG------------------LILGLFAQFTIAYAIDRSESAV
MAB_0651c|M.abscessus_ATCC_199 IG-SLLIIGG------------------VIFGFFAQYAVFFAIDRGLGPV
MSMEG_5268|M.smegmatis_MC2_155 VVAAILLFPGFFVPGYLFVGVPVLLIASAIIAVLFLFTTVAVLDRNLSGV
Mvan_4673|M.vanbaalenii_PYR-1 VGLFLCIVPG------------------VIASFLLMFTTIAVLDRNLAPM
Mflv_2044|M.gilvum_PYR-GCK IGYVLCIVPG------------------LIVSIFALFTTVFIVDRNLSAI
TH_4441|M.thermoresistible__bu IGTALCVVPG------------------LIVSLFAMFTTVAVVDRNLSPI
MLBr_02395|M.leprae_Br4923 ISEATGQPIG--------------------FGMSVVRQLAHFVDAVICCI
: * .. :* :
Mb2590|M.bovis_AF2122/97 DSVKASIETVGSNIGGSVLSWLAQLTAVLVG----ELLCFVGMLIGIPVA
Rv2560|M.tuberculosis_H37Rv DSVKASIETVGSNIGGSVLSWLAQLTAVLVG----ELLCFVGMLIGIPVA
MMAR_2166|M.marinum_M DSIKSSFATVRAELGNTALSWLVQYAVVLAG----ELLCFVGMLVGVPVA
MUL_1750|M.ulcerans_Agy99 DSIKSSCATVRAELGNTALSWLVQYAVVLAG----ELLCFVGMLVGVPVA
MAV_3436|M.avium_104 KALSSSFSTVTANLANALLVWLAEFALVVVG----ALACGVGLLLAAPVA
MAB_0651c|M.abscessus_ATCC_199 DALKASFQLVKDNLGQALVVFLITLGVAFGGFALTFITCGLGGIIAYPAA
MSMEG_5268|M.smegmatis_MC2_155 DAVKTSFELSKANFGTVFITAVVIFLLLLAG----AIACGIGLLVAYPLV
Mvan_4673|M.vanbaalenii_PYR-1 DAIKSSFETSKNNVGPVLLTWLASVAVVFVG----ALLCGVGLLVAAPLA
Mflv_2044|M.gilvum_PYR-GCK DGIKASIAVAKANFLQVFLTWLIFNVLISVG----SFVCYIGLIVTVPLA
TH_4441|M.thermoresistible__bu DGILASVDVVKKNFGPAILVWLASGVLIFLG----SLLCGVGLLITGPLA
MLBr_02395|M.leprae_Br4923 GFLFPLWDSKRQTLADKIMTTVCLPI------------------------
: . . : :
Mb2590|M.bovis_AF2122/97 ----ALIHVYTYRKLSGGQVVEAV--RPAPPVGWPPGP--------QLA
Rv2560|M.tuberculosis_H37Rv ----ALIHVYTYRKLSGGQVVEAV--RPAPPVGWPPGP--------QLA
MMAR_2166|M.marinum_M ----SLIQTYTWRKLSGGQVVELS--QPGPPAGLPPGPPPGAPPGPQFG
MUL_1750|M.ulcerans_Agy99 ----SLIQTYTWRKLSGGQVVELS--QPGPPAGLPPGPPPGAPPGPQFG
MAV_3436|M.avium_104 ----ALVGIYAYRKLSGGQVAPPPGYQPGPPPGYQPGPPPGAPPA----
MAB_0651c|M.abscessus_ATCC_199 GALTGLIHVYTYRRLTGGTIAPAPV------------------------
MSMEG_5268|M.smegmatis_MC2_155 ----ALIEVYAFRRLTGAPIAPPTQ------------------------
Mvan_4673|M.vanbaalenii_PYR-1 ----ALILVYAYRTLNGGFVAPAVP------------------------
Mflv_2044|M.gilvum_PYR-GCK ----VLYMVYAYRTLTGGYVAPATP------------------------
TH_4441|M.thermoresistible__bu ----GLLVVCTFRRLTGAPVAPQAV------------------------
MLBr_02395|M.leprae_Br4923 -------------------------------------------------