For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSVERRKPPYRTAGLAALAIMALLLVLVGQSFRGAFDPKVRLTLLTARAGLVLDPGAKVTYNGVPVGRVA EVGTAARTGTGVRARVMLDVDPRYVPLLPANVEASVTATTVFGNKYVALRSPEHPSAQRVSPATPIDATG VTTEFNTLFETIMRITEQVDPIALNQTLSAAAQALAGLGDRFGASLLDGNAILDDLNARIPPLRRDVAGL ADLAAIYADSAPDLFTGLDDAARGAATLNSQRGEVDAALIAAVGFAGTTSDILERGGPYLLRGAADLLPT SRLLDDYRAMIFCTIRNYHDVAPELDRALGGDNGYSLSAAGTPSSIGAGNVYVYPDNLPRVNAKGGPEGR PGCWQKITRELWPAPYLVMDTGLSVAPYNHVELGSPIFVDYVWGRQIGEPTINP
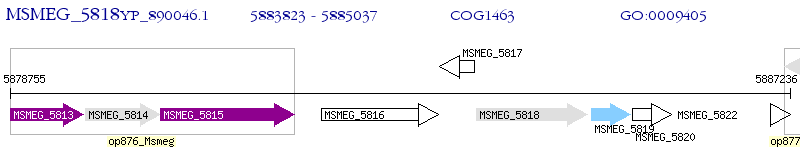
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5818 | - | - | 100% (404) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_0134 | - | e-149 | 63.66% (399) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_6540 | - | e-142 | 60.25% (400) | virulence factor Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0175 | mce1A | 1e-134 | 53.03% (445) | MCE-family protein MCE1A |
| M. gilvum PYR-GCK | Mflv_0711 | - | 1e-143 | 60.50% (400) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv0169 | mce1A | 1e-134 | 53.03% (445) | MCE-family protein MCE1A |
| M. leprae Br4923 | MLBr_02589 | mce1A | 1e-130 | 53.81% (433) | putative cell invasion protein |
| M. abscessus ATCC 19977 | MAB_4153c | - | 8e-50 | 31.27% (387) | MCE-family protein |
| M. marinum M | MMAR_4883 | mce2A | 1e-136 | 58.25% (400) | MCE-family protein Mce2A |
| M. avium 104 | MAV_4551 | - | 1e-135 | 61.38% (391) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_0154 | - | 1e-139 | 59.65% (399) | virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_0415 | mce2A | 1e-132 | 57.25% (400) | MCE-family protein Mce2A |
| M. vanbaalenii PYR-1 | Mvan_0134 | - | 1e-142 | 60.00% (400) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0175|M.bovis_AF2122/97 MTTPGK-LNKARVPPYKTAGLGLVLVFALVVALVYLQFRGEFTPKTQLTM
Rv0169|M.tuberculosis_H37Rv MTTPGK-LNKARVPPYKTAGLGLVLVFALVVALVYLQFRGEFTPKTQLTM
MLBr_02589|M.leprae_Br4923 MTSSTK-INASRKPPYKLAGVGLLVVAVVIFGLVYGQFRGNFTPKSKLTM
MAV_4551|M.avium_104 MATPVP-QRGRRAWVY-VEGVLLLLVCALVLALVYLQFRGDFTPKTKLTM
Mflv_0711|M.gilvum_PYR-GCK MTAP---MNTKRQPPYKIAGVVLALVTILILVLVYLQFRGDFLPRTQLTM
Mvan_0134|M.vanbaalenii_PYR-1 MTVPQAALNTRRQPPYKLAGLVLAILTVIAVTLVYLQFRGDFMARTQLTM
TH_0154|M.thermoresistible__bu MTAP---LNAPRTPPYKIVGVVLLLVVALVVTLVWMQFRGAFTPRTQLTL
MSMEG_5818|M.smegmatis_MC2_155 MSVE------RRKPPYRTAGLAALAIMALLLVLVGQSFRGAFDPKVRLTL
MMAR_4883|M.marinum_M -------MDPRSRPPYKIIGLAALMVLVFFGAMLYWQFRGTFIPKTRLTM
MUL_0415|M.ulcerans_Agy99 -------MDPRSRPPCKIIGLAALMVLVFFGAMLYWQFRGTFIPKTRLTM
MAB_4153c|M.abscessus_ATCC_199 ------MSDTSTRRAVRLAGTVLASVLVLLTVLTYLAYNDAFADTKPITV
* : . : :.. * :*:
Mb0175|M.bovis_AF2122/97 LSARAGLVMDPGSKVTYNGVEIGRVDTISEVTRDGE-SAAKFILDVDPRY
Rv0169|M.tuberculosis_H37Rv LSARAGLVMDPGSKVTYNGVEIGRVDTISEVTRDGE-SAAKFILDVDPRY
MLBr_02589|M.leprae_Br4923 LADRAGLVMDRGSKITYNGVKIGRVADISETVRNGR-PVAEFTLDVYPRY
MAV_4551|M.avium_104 VATRAGLVMETGSKVTYNGVAIGRVADISEIERDGA-PAAKLVLDVDPRY
Mflv_0711|M.gilvum_PYR-GCK ISARSGLSLDSGSKVTFNGVEIGKVSEVEETTVGGE-PRAKILLEVNPKY
Mvan_0134|M.vanbaalenii_PYR-1 ISARSGLSMDPGAKVTYNGVEIGRVGNVEETSVDGE-PRAKIILDVNPKY
TH_0154|M.thermoresistible__bu LADRAGLVMDPGSKVTFNGVEIGRVAQIHADTSDDR-TRANIVLDVDPKY
MSMEG_5818|M.smegmatis_MC2_155 LTARAGLVLDPGAKVTYNGVPVGRVAEVGTAARTGTGVRARVMLDVDPRY
MMAR_4883|M.marinum_M IAPRAGLVTDPGAKVTFNGVQIGRVSSITEVTHDGA-PAARFALDVTPKY
MUL_0415|M.ulcerans_Agy99 IAPRAGLVTDPGAKVTFNGVQIGRVSSITEVTHDGA-PAARFALDVTPKY
MAB_4153c|M.abscessus_ATCC_199 VSPRAGLVMEEGGKVKFHGLQIGKVTSISYAN-----DQAKLELAVNAKD
:: *:** : *.*:.::*: :*:* : *.. * * .:
Mb0175|M.bovis_AF2122/97 IHLIPANVNADIKATTVFGGKYVSLTTPKNPTKRRITPKDVIDVRSVTTE
Rv0169|M.tuberculosis_H37Rv IHLIPANVNADIKATTVFGGKYVSLTTPKNPTKRRITPKDVIDVRSVTTE
MLBr_02589|M.leprae_Br4923 LSLIPANVNADIKATTVFGGKYVSLTTPEHPSQKRLTPQTVIDARSVTTE
MAV_4551|M.avium_104 VNLIPANVVATIEAATLFGNKYVSLSAPENPSRQRISPRDVIDARSVTTE
Mflv_0711|M.gilvum_PYR-GCK LKLIPRNVNADISATTVFGNKYVSFTSPENPQSERISASDVIDVTSVTTE
Mvan_0134|M.vanbaalenii_PYR-1 LDLIPSNVNADISATTVFGNKYVSFTTPEDPRPERISASDVIDVTSVTTE
TH_0154|M.thermoresistible__bu VDLIPSNADVEIKASTVFGNKYVAFASPPDPSLTPITSAATIDVTKVTTE
MSMEG_5818|M.smegmatis_MC2_155 VPLLPANVEASVTATTVFGNKYVALRSPEHPSAQRVSPATPIDATGVTTE
MMAR_4883|M.marinum_M IGSIPANVSATIKATTVFGNKYVSLTSPTDPAPQHITPASVIYATSVTTE
MUL_0415|M.ulcerans_Agy99 IGSIPANVSATIKATTVFGNKYVSLTSPTDPAPQHITPASVIYATSVTTE
MAB_4153c|M.abscessus_ATCC_199 LRLIPSNATVRIAGTTVFGAKSVEFLAPQKPSDSSLRPGATVTAEAVALE
: :* *. . : .:*:** * * : :* .* : . : . *: *
Mb0175|M.bovis_AF2122/97 INTLFQTLTSIAEKVDPVKLNLTLSAAAEALTGLGDKFGESIVNANTVLD
Rv0169|M.tuberculosis_H37Rv INTLFQTLTSIAEKVDPVKLNLTLSAAAEALTGLGDKFGESIVNANTVLD
MLBr_02589|M.leprae_Br4923 INTLFQTITLIAEKVDPIKLNLTLSAAAQSLAGLGERFGQSIVNGNSVLD
MAV_4551|M.avium_104 FNTLFETVTSIAEKVSPIELNATLSALAQALDGLGGKFGESIVNGNQILA
Mflv_0711|M.gilvum_PYR-GCK FNTLFETVVEIAEQVDPIKLNQTLTATAQALDGLGDRFGESIINGNQILA
Mvan_0134|M.vanbaalenii_PYR-1 FNTLFETVVSLADQVDPIKLNQTLTATAQALDGLGDRFGESIINGNQILS
TH_0154|M.thermoresistible__bu FNTLFQTVVDIARQVDPIKLNQTLTATAQALDGLGDRFGESIIDGNAILD
MSMEG_5818|M.smegmatis_MC2_155 FNTLFETIMRITEQVDPIALNQTLSAAAQALAGLGDRFGASLLDGNAILD
MMAR_4883|M.marinum_M FNTLFENVTAISEKVDPVKLNLTLSAAAQALSGLGDRLGTALVNGNAILD
MUL_0415|M.ulcerans_Agy99 FNTLFENVTAISEKVDPDKLNLTLNAAAQALSGLGDRLGTALVNGNAILD
MAB_4153c|M.abscessus_ATCC_199 VNTVFENLTRLLGKIDPVNLNATLTAVGEGLRGNGDNFGQGLVELDQYLD
.**:*:.: : ::.* ** **.* .:.* * * .:* .::: : *
Mb0175|M.bovis_AF2122/97 DLNSRMPQSRHDIQQLAALGDVYADAAPDLFDFLDSSVTTARTINAQQAE
Rv0169|M.tuberculosis_H37Rv DLNSRMPQSRHDIQQLAALGDVYADAAPDLFDFLDSSVTTARTINAQQAE
MLBr_02589|M.leprae_Br4923 DVNSQLPQARHDIQQLASLGDTYANSASDFFDFLNNSIVTSRTINQQQKD
MAV_4551|M.avium_104 QLNPRMPQIRYDVRRLADLAVAYTKAAPDLLDFLSNAVTTARTLTRQQGD
Mflv_0711|M.gilvum_PYR-GCK EINPQMPQIRRDNQLLADFGEVYSNAAPDLFDGLQNAVTTARTLNEQQGD
Mvan_0134|M.vanbaalenii_PYR-1 EINPQMPQIRRDNQLLADLGDVYADAAPDLFDGLQNAVTTARTLNEQQGD
TH_0154|M.thermoresistible__bu DLNPQMPQIRYDVQRLADLNAVYAAAAPDLFDGLENAVTTAQTLYQERGN
MSMEG_5818|M.smegmatis_MC2_155 DLNARIPPLRRDVAGLADLAAIYADSAPDLFTGLDDAARGAATLNSQRGE
MMAR_4883|M.marinum_M DVNPLMPQVHRDITGLAALGDIYADASTDLWDAVNSTVTVARTVNQRRSD
MUL_0415|M.ulcerans_Agy99 DVNPLMPQVHRDIRGLAALGDIYADASADLWDAVNSTVTVARTVNQRRSD
MAB_4153c|M.abscessus_ATCC_199 QLNPKLPALQQDFRQAGQVGQIYGDAAPDIVRILDNLPALSRTVVDQQQD
::*. :* : * . . * ::.*: :.. : *: .: :
Mb0175|M.bovis_AF2122/97 LDSALLAAAGFGNTTADVFDRGGPYLQRGVADLVPTATLLDTYSPELFCT
Rv0169|M.tuberculosis_H37Rv LDSALLAAAGFGNTTADVFDRGGPYLQRGVADLVPTATLLDTYSPELFCT
MLBr_02589|M.leprae_Br4923 LDQVLLAAVGFGNTGADIFSRSGPYLARGAADLVPTAQLLDTYSPAIFCT
MAV_4551|M.avium_104 LDAALLAAVGVGHEGEDIFARGGPYLARGAADLVPTAELLDTYSPELFCM
Mflv_0711|M.gilvum_PYR-GCK VDQALMAAVGFGNTGGDIFERGGPYLVRGARDLVPTSALLDKYSPSFFCM
Mvan_0134|M.vanbaalenii_PYR-1 VDQALMAAVGFGNTGGDVFERGGPYLARGAEDLVPTTALLDKYSPSFFCM
TH_0154|M.thermoresistible__bu LDQTLLASIGFGNTAGEVFERGGPYLVRGSEDLVTVTEILDYHSPALFCM
MSMEG_5818|M.smegmatis_MC2_155 VDAALIAAVGFAGTTSDILERGGPYLLRGAADLLPTSRLLDDYRAMIFCT
MMAR_4883|M.marinum_M LDAALLASVGLANNGSEVFERGGPYLTRGAADLVPTGQLLDEYSPEIFCT
MUL_0415|M.ulcerans_Agy99 LDAALLASVGLANNGSEVFERGGPYLTRGAADLVPTGQLLDEYSPEIFRT
MAB_4153c|M.abscessus_ATCC_199 LKATLLAAIGLGNNGYETLAPAEKDLTAALQRLRAPATLLGRYSPEIPCT
:. .*:*: *.. : : . * . * . :*. : . :
Mb0175|M.bovis_AF2122/97 IRNFYDADPLAKAAAGGGNGYSLRTNSEILSGIGISLLSPLALATNGAAI
Rv0169|M.tuberculosis_H37Rv IRNFYDADPLAKAASGGGNGYSLRTNSEILSGIGISLLSPLALATNGAAI
MLBr_02589|M.leprae_Br4923 LRNYHDIEPKVTSFLGG-NGYSINDHVELLSGLGFLLNPTSAISIIGAGV
MAV_4551|M.avium_104 IRNFHDAAPEVAKAAGG-NGYSLAA-------------------------
Mflv_0711|M.gilvum_PYR-GCK MRNYHDVEPKIADALGG-NGYSLKTHS-----------------------
Mvan_0134|M.vanbaalenii_PYR-1 IRNYHDVEPKIADALGG-NGYSLKTHS-----------------------
TH_0154|M.thermoresistible__bu IRNYHDVAPKVAASLGG-NGYSLRQHA-----------------------
MSMEG_5818|M.smegmatis_MC2_155 IRNYHDVAPELDRALGGDNGYSLSAAG-----------------------
MMAR_4883|M.marinum_M IRNYDEVAPRIRNALGF-NGYSLGAAS-----------------------
MUL_0415|M.ulcerans_Agy99 IRNYDEVAPRIRNALGF-NGYSLGAAP-----------------------
MAB_4153c|M.abscessus_ATCC_199 LQAISKALVTFTPLIGGVRPGLFLTNS-----------------------
:: . * . :
Mb0175|M.bovis_AF2122/97 GIGLVAGLIASPLAVAANLAGALPGIVGGAPNPYTYPENLPRVNARGGPG
Rv0169|M.tuberculosis_H37Rv GIGLVAGLIAPPLAVAANLAGALPGIVGGAPNPYTYPENLPRVNARGGPG
MLBr_02589|M.leprae_Br4923 ---LSQGLLAP-------LG--LAGVVNGAANPYIYPENLPRVNARGGPG
MAV_4551|M.avium_104 -----------------------AGSIVGAPNPYVYPDNLPRVNAHGGPG
Mflv_0711|M.gilvum_PYR-GCK -----------------------QVLGLGAGNAYVYPDNLPRVNAKGGPG
Mvan_0134|M.vanbaalenii_PYR-1 -----------------------EVLGLGAGNAYVYPDNLPRVNAKGGPG
TH_0154|M.thermoresistible__bu -----------------------FLVGP--GNPYVYPDNLPRVNATGGPG
MSMEG_5818|M.smegmatis_MC2_155 -----------------------TPSSIGAGNVYVYPDNLPRVNAKGGPE
MMAR_4883|M.marinum_M -----------------------SGAISGAPNPYIYPENLPRVNARGGPG
MUL_0415|M.ulcerans_Agy99 -----------------------SGAISGAPNPYIYPENLPRVNARGGPG
MAB_4153c|M.abscessus_ATCC_199 --------------------------FPLGAPVYTYPDSLPIVNASGGPN
* **:.** *** ***
Mb0175|M.bovis_AF2122/97 GA--PGCWQPITRDLWP-APYLVMDTG-ASLAPYNHMEVGSPYAVEYVWG
Rv0169|M.tuberculosis_H37Rv GA--PGCWQPITRDLWP-APYLVMDTG-ASLAPYNHMEVGSPYAVEYVWG
MLBr_02589|M.leprae_Br4923 GA--PGCWQHITRDLWP-TPELVMDTG-NSIAPYNHLDSGSPYAIEYVWG
MAV_4551|M.avium_104 GR--PGCWQKITRDLWP-APYLVMDTG-ASLAPYNHLEIGQPLATEFVWG
Mflv_0711|M.gilvum_PYR-GCK GR--PGCWQPITRDLWP-APFLVLDTG-ASVAPYNHMELGQPLFTEYVWG
Mvan_0134|M.vanbaalenii_PYR-1 GR--PGCWQPITRDLWP-APFLVLDTG-ASIAPYNHMELGQPLLTEYVWG
TH_0154|M.thermoresistible__bu GR--PGCWQPITRDLWP-APYLVMDTG-ASLAPYNHIELGKPFAIEYVWG
MSMEG_5818|M.smegmatis_MC2_155 GR--PGCWQKITRELWP-APYLVMDTG-LSVAPYNHVELGSPIFVDYVWG
MMAR_4883|M.marinum_M GR--PGCWQTITRDLWP-APYLVMDVG-ANMAPYNHLEIGQPLLNEYVWG
MUL_0415|M.ulcerans_Agy99 GR--PGCWQTITRDLWP-APYLVMNVG-ANMAPYNHLEIGQPLLNEYVWG
MAB_4153c|M.abscessus_ATCC_199 CRGLPNIPTKMQNGSWFRSPFLVTDNAYIPYKPLEELQVNAPDTLQYLYN
*. : . * :* ** : . * :.:: . * ::::.
Mb0175|M.bovis_AF2122/97 RQVGDNTINP
Rv0169|M.tuberculosis_H37Rv RQVGDNTINP
MLBr_02589|M.leprae_Br4923 RQVGDNTINP
MAV_4551|M.avium_104 RQYGENTINP
Mflv_0711|M.gilvum_PYR-GCK RQVGEHTINP
Mvan_0134|M.vanbaalenii_PYR-1 RQVGEHTINP
TH_0154|M.thermoresistible__bu RQIGEYTINP
MSMEG_5818|M.smegmatis_MC2_155 RQIGEPTINP
MMAR_4883|M.marinum_M RQFGENTINP
MUL_0415|M.ulcerans_Agy99 RQFGENTINP
MAB_4153c|M.abscessus_ATCC_199 GAFAKRSEF-
.. :