For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VASSAALQDRLGQRFKHWRYDPPYKTACAGMVILVVTVLVLTWMQFRGDFEDKAEITLFASRSGLSMDPG SKVTFNGVPIGRLKGVDVVEVDDNPEARLTLDIDPKYLDLIPANAVAELRATTVFGNKYVSFTAPEHPSA ERLSASTPIRAQGVTTEFNTLFETITAISEQVDPIKLNETLTAAAQALDGLGDKFGRSIVDGNAILADVN PRMPQIRRDITGLANLGEVYADAAPDLFDGLDNAVTTARTLNDQRHDLDQALVAAVGFGNTGGDIFERGG PYLVRGNQDLLPVAEMFDRNSPALYCTIKNYHDAAPKFAAQTNNGYSFQLNDTFVGAGNPYVYPDNLPRV NARGGPEGRPGCWQPITHDLWPAPYLVLDTGASIAPYNHFELGQPMFIEYVWGRQVGENTINP
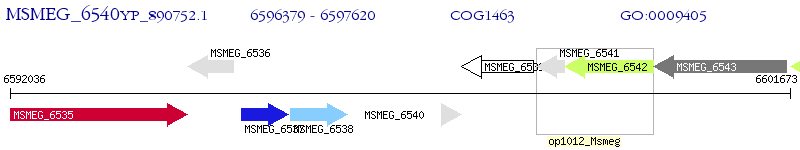
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6540 | - | - | 100% (413) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_0134 | - | 0.0 | 83.97% (393) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_5818 | - | e-142 | 60.25% (400) | virulence factor Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0604 | mce2A | 1e-142 | 63.05% (387) | MCE-family protein MCE2A |
| M. gilvum PYR-GCK | Mflv_0711 | - | 1e-157 | 66.08% (395) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv0589 | mce2A | 1e-142 | 63.05% (387) | MCE-family protein MCE2A |
| M. leprae Br4923 | MLBr_02589 | mce1A | 1e-136 | 55.24% (429) | putative cell invasion protein |
| M. abscessus ATCC 19977 | MAB_4153c | - | 5e-59 | 35.17% (381) | MCE-family protein |
| M. marinum M | MMAR_0412 | mce1A | 1e-141 | 56.97% (423) | MCE-family protein Mce1A |
| M. avium 104 | MAV_4551 | - | 1e-142 | 63.57% (387) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_0154 | - | 1e-153 | 64.38% (393) | virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_1062 | mce1A | 1e-140 | 56.74% (423) | MCE-family protein Mce1A |
| M. vanbaalenii PYR-1 | Mvan_0134 | - | 1e-159 | 66.58% (398) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0711|M.gilvum_PYR-GCK MTAP---MNTKRQ--------PPYKIAGVVLALVTILILVLVYLQFRGDF
Mvan_0134|M.vanbaalenii_PYR-1 MTVPQAALNTRRQ--------PPYKLAGLVLAILTVIAVTLVYLQFRGDF
TH_0154|M.thermoresistible__bu MTAP---LNAPRT--------PPYKIVGVVLLLVVALVVTLVWMQFRGAF
MSMEG_6540|M.smegmatis_MC2_155 MASSAALQDRLGQRFKHWRYDPPYKTACAGMVILVVTVLVLTWMQFRGDF
Mb0604|M.bovis_AF2122/97 MPTLVTRKNRRAW----------LYVEGVVLLLVGALVLVLVYKQFRGEF
Rv0589|M.tuberculosis_H37Rv MPTLVTRKNRRAW----------LYVEGVVLLLVGALVLVLVYKQFRGEF
MAV_4551|M.avium_104 MATPVPQRGRRAW----------VYVEGVLLLLVCALVLALVYLQFRGDF
MMAR_0412|M.marinum_M MTKPGKLNKVNEP---------PYKIAGIALFLVGLLILSLVYLQFRGDF
MUL_1062|M.ulcerans_Agy99 MTKPGKLNKVNEP---------PYKIAGIALFLVGLLILSLVYLQFRGDF
MLBr_02589|M.leprae_Br4923 MTSSTKINASRKP---------PYKLAGVGLLVVAVVIFGLVYGQFRGNF
MAB_4153c|M.abscessus_ATCC_199 MSDTSTRR--------------AVRLAGTVLASVLVLLTVLTYLAYNDAF
*. : : *.: :.. *
Mflv_0711|M.gilvum_PYR-GCK LPRTQLTMISARSGLSLDSGSKVTFNGVEIGKVSEVEETTVGGEPRAKIL
Mvan_0134|M.vanbaalenii_PYR-1 MARTQLTMISARSGLSMDPGAKVTYNGVEIGRVGNVEETSVDGEPRAKII
TH_0154|M.thermoresistible__bu TPRTQLTLLADRAGLVMDPGSKVTFNGVEIGRVAQIHADTSDDRTRANIV
MSMEG_6540|M.smegmatis_MC2_155 EDKAEITLFASRSGLSMDPGSKVTFNGVPIGRLKGVDVVEVDDNPEARLT
Mb0604|M.bovis_AF2122/97 TPKTELTMVASRAGLVMEAGSKVTYNGVEIGRVGSISEIERDGRPAAKLV
Rv0589|M.tuberculosis_H37Rv TPKTELTMVAFRAGLVMEAGSKVTYNGVEIGRVGSISEIERDGRPAAKLV
MAV_4551|M.avium_104 TPKTKLTMVATRAGLVMETGSKVTYNGVAIGRVADISEIERDGAPAAKLV
MMAR_0412|M.marinum_M DRKVKLTMFSERAGLVMDPGSKVTYNGVQIGRVDTISEITRDGKPAAKFV
MUL_1062|M.ulcerans_Agy99 DRKVKLTMFSERAGLVMDPGSKVTYNGVQIGRVDTISEITRDGKPAAKFV
MLBr_02589|M.leprae_Br4923 TPKSKLTMLADRAGLVMDRGSKITYNGVKIGRVADISETVRNGRPVAEFT
MAB_4153c|M.abscessus_ATCC_199 ADTKPITVVSPRAGLVMEEGGKVKFHGLQIGKVTSISYAN----DQAKLE
:*:.: *:** :: *.*:.::*: **:: : *.:
Mflv_0711|M.gilvum_PYR-GCK LEVNPKYLKLIPRNVNADISATTVFGNKYVSFTSPENPQSERISASDVID
Mvan_0134|M.vanbaalenii_PYR-1 LDVNPKYLDLIPSNVNADISATTVFGNKYVSFTTPEDPRPERISASDVID
TH_0154|M.thermoresistible__bu LDVDPKYVDLIPSNADVEIKASTVFGNKYVAFASPPDPSLTPITSAATID
MSMEG_6540|M.smegmatis_MC2_155 LDIDPKYLDLIPANAVAELRATTVFGNKYVSFTAPEHPSAERLSASTPIR
Mb0604|M.bovis_AF2122/97 LDVNPRYISLIPVNVVADIEAATLFGNKYVALSAPKIPQQQRISSHDVID
Rv0589|M.tuberculosis_H37Rv LDVNPRYISLIPVNVVADIEAATLFGNKYVALSAPKIPQQQRISSHDVID
MAV_4551|M.avium_104 LDVDPRYVNLIPANVVATIEAATLFGNKYVSLSAPENPSRQRISPRDVID
MMAR_0412|M.marinum_M LNVYPRYLPLIPENVDAQIKATTVFGGKYVSLTTPEHPSNKKLSAHSVIN
MUL_1062|M.ulcerans_Agy99 LNVYPRYLPLIPENVDAQIKATTVFGGKYVSLTTPEHPSNKKLSAHSVIN
MLBr_02589|M.leprae_Br4923 LDVYPRYLSLIPANVNADIKATTVFGGKYVSLTTPEHPSQKRLTPQTVID
MAB_4153c|M.abscessus_ATCC_199 LAVNAKDLRLIPSNATVRIAGTTVFGAKSVEFLAPQKPSDSSLRPGATVT
* : .: : *** *. . : .:*:** * * : :* * : . :
Mflv_0711|M.gilvum_PYR-GCK VTSVTTEFNTLFETVVEIAEQVDPIKLNQTLTATAQALDGLGDRFGESII
Mvan_0134|M.vanbaalenii_PYR-1 VTSVTTEFNTLFETVVSLADQVDPIKLNQTLTATAQALDGLGDRFGESII
TH_0154|M.thermoresistible__bu VTKVTTEFNTLFQTVVDIARQVDPIKLNQTLTATAQALDGLGDRFGESII
MSMEG_6540|M.smegmatis_MC2_155 AQGVTTEFNTLFETITAISEQVDPIKLNETLTAAAQALDGLGDKFGRSIV
Mb0604|M.bovis_AF2122/97 VGSVTTEFNTLFETITSIAEKVDPIELNATLSAVAQAPDGLGGKFGESIV
Rv0589|M.tuberculosis_H37Rv VGSVTTEFNTLFETITSIAEKVDPIELNATLSAVAQALDGLGGKFGESIV
MAV_4551|M.avium_104 ARSVTTEFNTLFETVTSIAEKVSPIELNATLSALAQALDGLGGKFGESIV
MMAR_0412|M.marinum_M ASAVTTEINTLFETLTSISEKVDPVKLNLTLSAAAEALTGLGNRFGEAIA
MUL_1062|M.ulcerans_Agy99 ASAVTIEINTLFETLTSISEKVDPVKLNLTLSAAAEALTGLGNRFGEAIA
MLBr_02589|M.leprae_Br4923 ARSVTTEINTLFQTITLIAEKVDPIKLNLTLSAAAQSLAGLGERFGQSIV
MAB_4153c|M.abscessus_ATCC_199 AEAVALEVNTVFENLTRLLGKIDPVNLNATLTAVGEGLRGNGDNFGQGLV
. *: *.**:*:.:. : ::.*::** **:* .:. * * .**..:
Mflv_0711|M.gilvum_PYR-GCK NGNQILAEINPQMPQIRRDNQLLADFGEVYSNAAPDLFDGLQNAVTTART
Mvan_0134|M.vanbaalenii_PYR-1 NGNQILSEINPQMPQIRRDNQLLADLGDVYADAAPDLFDGLQNAVTTART
TH_0154|M.thermoresistible__bu DGNAILDDLNPQMPQIRYDVQRLADLNAVYAAAAPDLFDGLENAVTTAQT
MSMEG_6540|M.smegmatis_MC2_155 DGNAILADVNPRMPQIRRDITGLANLGEVYADAAPDLFDGLDNAVTTART
Mb0604|M.bovis_AF2122/97 NGNQILAQLNPRLPQLGYDVRRLADLGEVYVDASPDLWSFLQNALTTART
Rv0589|M.tuberculosis_H37Rv NGNQILAQLNPRLPQLGYDVRRLADLGEVYVDASPDLWSFLQNALTTART
MAV_4551|M.avium_104 NGNQILAQLNPRMPQIRYDVRRLADLAVAYTKAAPDLLDFLSNAVTTART
MMAR_0412|M.marinum_M NGNEILDDINPQMPQARRDIQQLAALGDVYADAAPDLFDFLNNSVPTART
MUL_1062|M.ulcerans_Agy99 NGNEILDDINPQMPQARRDIQQLAALGDVYADAAPDLFDFLNNSVPTART
MLBr_02589|M.leprae_Br4923 NGNSVLDDVNSQLPQARHDIQQLASLGDTYANSASDFFDFLNNSIVTSRT
MAB_4153c|M.abscessus_ATCC_199 ELDQYLDQLNPKLPALQQDFRQAGQVGQIYGDAAPDIVRILDNLPALSRT
: : * ::*.::* * . . * ::.*: *.* ::*
Mflv_0711|M.gilvum_PYR-GCK LNEQQGDVDQALMAAVGFGNTGGDIFERGGPYLVRGARDLVPTSALLDKY
Mvan_0134|M.vanbaalenii_PYR-1 LNEQQGDVDQALMAAVGFGNTGGDVFERGGPYLARGAEDLVPTTALLDKY
TH_0154|M.thermoresistible__bu LYQERGNLDQTLLASIGFGNTAGEVFERGGPYLVRGSEDLVTVTEILDYH
MSMEG_6540|M.smegmatis_MC2_155 LNDQRHDLDQALVAAVGFGNTGGDIFERGGPYLVRGNQDLLPVAEMFDRN
Mb0604|M.bovis_AF2122/97 LTSQQRDLDAALLAATGAGNTGEDVFARGGPYLARAAADLVPTATLLDTY
Rv0589|M.tuberculosis_H37Rv LTSQQRDLDAALLAATGAGNTGEDVFARGGPYLARAAADLVPTATLLDTY
MAV_4551|M.avium_104 LTRQQGDLDAALLAAVGVGHEGEDIFARGGPYLARGAADLVPTAELLDTY
MMAR_0412|M.marinum_M INAQQHDLDAALLAAVGFGNTGADVFERGGPYLARGAADLVPSAQLLDTY
MUL_1062|M.ulcerans_Agy99 INAQQHDLDAALLAAVGFGNTGADVFERGGPYLARGAADLVPSAQLLDTY
MLBr_02589|M.leprae_Br4923 INQQQKDLDQVLLAAVGFGNTGADIFSRSGPYLARGAADLVPTAQLLDTY
MAB_4153c|M.abscessus_ATCC_199 VVDQQQDLKATLLAAIGLGNNGYETLAPAEKDLTAALQRLRAPATLLGRY
: :: ::. .*:*: * *: . : : . *. . * . : ::.
Mflv_0711|M.gilvum_PYR-GCK SPSFFCMMRNYHDVEPKIADALGGNGYSLKTHSQVLGLG-----------
Mvan_0134|M.vanbaalenii_PYR-1 SPSFFCMIRNYHDVEPKIADALGGNGYSLKTHSEVLGLG-----------
TH_0154|M.thermoresistible__bu SPALFCMIRNYHDVAPKVAASLGGNGYSLRQHAFLVGP------------
MSMEG_6540|M.smegmatis_MC2_155 SPALYCTIKNYHDAAPKFAAQTN-NGYSFQLNDTFVGA------------
Mb0604|M.bovis_AF2122/97 SPELFCMIRNFHDAAPKVADAVGGNGYSLAAAGTILGAP-----------
Rv0589|M.tuberculosis_H37Rv SPELFCMIRNFHDAAPKVADAVGGNGYSLAAAGTILGAP-----------
MAV_4551|M.avium_104 SPELFCMIRNFHDAAPEVAKAAGGNGYSLAAAGSIVGAP-----------
MMAR_0412|M.marinum_M SPELFCTLRNYHDVEPQAYKFLGGNGYSLNSHTTPLSALGMMLNPASLVP
MUL_1062|M.ulcerans_Agy99 SPELFCTLRNYHDVEPQAYKFLGGNGYSLNSHTTPLSALGMMLNPASLVP
MLBr_02589|M.leprae_Br4923 SPAIFCTLRNYHDIEPKVTSFLGGNGYSINDHVELLSGLGFLLNPTSAIS
MAB_4153c|M.abscessus_ATCC_199 SPEIPCTLQAISKALVTFTPLIGGVRPGLFLTNSFPLGAP----------
** : * :: . . .:
Mflv_0711|M.gilvum_PYR-GCK -----------------------AGNAYVYPDNLPRVNAKGGP--GGRPG
Mvan_0134|M.vanbaalenii_PYR-1 -----------------------AGNAYVYPDNLPRVNAKGGP--GGRPG
TH_0154|M.thermoresistible__bu ------------------------GNPYVYPDNLPRVNATGGP--GGRPG
MSMEG_6540|M.smegmatis_MC2_155 ------------------------GNPYVYPDNLPRVNARGGP--EGRPG
Mb0604|M.bovis_AF2122/97 -------------------------NPYVYPDNLPRVNAHGGP--GGRPG
Rv0589|M.tuberculosis_H37Rv -------------------------NPYVYPDNLPRVNAHGGP--GGRPG
MAV_4551|M.avium_104 -------------------------NPYVYPDNLPRVNAHGGP--GGRPG
MMAR_0412|M.marinum_M LL---ISQVGGLA----AGLVGGAPNPYIYPENLPRVNAHGGP--GGAPG
MUL_1062|M.ulcerans_Agy99 LL---ISQVGGLA----AGLVGGAPNPYIYPENLPRVNAHGGP--GGAPG
MLBr_02589|M.leprae_Br4923 IIGAGVLSQGLLAPLGLAGVVNGAANPYIYPENLPRVNARGGP--GGAPG
MAB_4153c|M.abscessus_ATCC_199 --------------------------VYTYPDSLPIVNASGGPNCRGLPN
* **:.** *** *** * *.
Mflv_0711|M.gilvum_PYR-GCK CWQPITRDLWP-APFLVLDTG-ASVAPYNHMELGQPLFTEYVWGRQVGEH
Mvan_0134|M.vanbaalenii_PYR-1 CWQPITRDLWP-APFLVLDTG-ASIAPYNHMELGQPLLTEYVWGRQVGEH
TH_0154|M.thermoresistible__bu CWQPITRDLWP-APYLVMDTG-ASLAPYNHIELGKPFAIEYVWGRQIGEY
MSMEG_6540|M.smegmatis_MC2_155 CWQPITHDLWP-APYLVLDTG-ASIAPYNHFELGQPMFIEYVWGRQVGEN
Mb0604|M.bovis_AF2122/97 CWQTITRELWP-APYLVMDTG-ASLAPYNHVELGQPMFTEYVWGRQYGEN
Rv0589|M.tuberculosis_H37Rv CWQTITRELWP-APYLVMDTG-ASLAPYNHVELGQPMFTEYVWGRQYGEN
MAV_4551|M.avium_104 CWQKITRDLWP-APYLVMDTG-ASLAPYNHLEIGQPLATEFVWGRQYGEN
MMAR_0412|M.marinum_M CWQSITHDFWP-APELVMDTG-NSLAPYNHLETGSPFVLEYVWGRQVGDY
MUL_1062|M.ulcerans_Agy99 CWQSITHDFWP-APELVMDTG-NSLAPYNHLETGSPFVLEYVWGRQVGDY
MLBr_02589|M.leprae_Br4923 CWQHITRDLWP-TPELVMDTG-NSIAPYNHLDSGSPYAIEYVWGRQVGDN
MAB_4153c|M.abscessus_ATCC_199 IPTKMQNGSWFRSPFLVTDNAYIPYKPLEELQVNAPDTLQYLYNGAFAKR
: . * :* ** *.. . * :..: . * ::::. ..
Mflv_0711|M.gilvum_PYR-GCK TINP
Mvan_0134|M.vanbaalenii_PYR-1 TINP
TH_0154|M.thermoresistible__bu TINP
MSMEG_6540|M.smegmatis_MC2_155 TINP
Mb0604|M.bovis_AF2122/97 TINP
Rv0589|M.tuberculosis_H37Rv TINP
MAV_4551|M.avium_104 TINP
MMAR_0412|M.marinum_M TINP
MUL_1062|M.ulcerans_Agy99 TINP
MLBr_02589|M.leprae_Br4923 TINP
MAB_4153c|M.abscessus_ATCC_199 SEF-
: