For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATPVPQRGRRAWVYVEGVLLLLVCALVLALVYLQFRGDFTPKTKLTMVATRAGLVMETGSKVTYNGVAIG RVADISEIERDGAPAAKLVLDVDPRYVNLIPANVVATIEAATLFGNKYVSLSAPENPSRQRISPRDVIDA RSVTTEFNTLFETVTSIAEKVSPIELNATLSALAQALDGLGGKFGESIVNGNQILAQLNPRMPQIRYDVR RLADLAVAYTKAAPDLLDFLSNAVTTARTLTRQQGDLDAALLAAVGVGHEGEDIFARGGPYLARGAADLV PTAELLDTYSPELFCMIRNFHDAAPEVAKAAGGNGYSLAAAGSIVGAPNPYVYPDNLPRVNAHGGPGGRP GCWQKITRDLWPAPYLVMDTGASLAPYNHLEIGQPLATEFVWGRQYGENTINP*
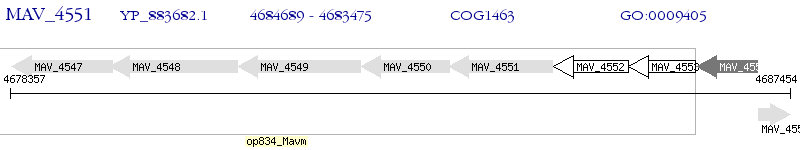
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4551 | - | - | 100% (404) | virulence factor Mce family protein |
| M. avium 104 | MAV_5015 | - | e-148 | 60.60% (434) | virulence factor Mce family protein |
| M. avium 104 | MAV_4126 | - | e-148 | 62.65% (415) | virulence factor Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0604 | mce2A | 0.0 | 82.59% (402) | MCE-family protein MCE2A |
| M. gilvum PYR-GCK | Mflv_0711 | - | 1e-156 | 67.01% (391) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv0589 | mce2A | 0.0 | 82.84% (402) | MCE-family protein MCE2A |
| M. leprae Br4923 | MLBr_02589 | mce1A | 1e-147 | 60.28% (423) | putative cell invasion protein |
| M. abscessus ATCC 19977 | MAB_4153c | - | 6e-61 | 36.27% (397) | MCE-family protein |
| M. marinum M | MMAR_0412 | mce1A | 1e-151 | 63.40% (418) | MCE-family protein Mce1A |
| M. smegmatis MC2 155 | MSMEG_0134 | - | 1e-148 | 65.63% (387) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_0154 | - | 1e-148 | 65.04% (389) | virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_1062 | mce1A | 1e-150 | 63.16% (418) | MCE-family protein Mce1A |
| M. vanbaalenii PYR-1 | Mvan_0134 | - | 1e-155 | 66.58% (389) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0412|M.marinum_M ------MTKP-GKLNKVNEPPYKIAGIALFLVGLLILSLVYLQFRGDFDR
MUL_1062|M.ulcerans_Agy99 ------MTKP-GKLNKVNEPPYKIAGIALFLVGLLILSLVYLQFRGDFDR
MLBr_02589|M.leprae_Br4923 ------MTSS-TKINASRKPPYKLAGVGLLVVAVVIFGLVYGQFRGNFTP
Mb0604|M.bovis_AF2122/97 ------MPTL--VTRKNRRAWLYVEGVVLLLVGALVLVLVYKQFRGEFTP
Rv0589|M.tuberculosis_H37Rv ------MPTL--VTRKNRRAWLYVEGVVLLLVGALVLVLVYKQFRGEFTP
MAV_4551|M.avium_104 ------MATP--VPQRGRRAWVYVEGVLLLLVCALVLALVYLQFRGDFTP
MSMEG_0134|M.smegmatis_MC2_155 MTEPPAPTAP---LNKPKTPPYKLAGLILGLVGVLVLALTWMQFRGQFED
TH_0154|M.thermoresistible__bu ------MTAP---LNAPRTPPYKIVGVVLLLVVALVVTLVWMQFRGAFTP
Mflv_0711|M.gilvum_PYR-GCK ------MTAP---MNTKRQPPYKIAGVVLALVTILILVLVYLQFRGDFLP
Mvan_0134|M.vanbaalenii_PYR-1 ------MTVPQAALNTRRQPPYKLAGLVLAILTVIAVTLVYLQFRGDFMA
MAB_4153c|M.abscessus_ATCC_199 ------------MSDTSTRRAVRLAGTVLASVLVLLTVLTYLAYNDAFAD
: * * : : *.: :.. *
MMAR_0412|M.marinum_M KVKLTMFSERAGLVMDPGSKVTYNGVQIGRVDTISEITRDGKPAAKFVLN
MUL_1062|M.ulcerans_Agy99 KVKLTMFSERAGLVMDPGSKVTYNGVQIGRVDTISEITRDGKPAAKFVLN
MLBr_02589|M.leprae_Br4923 KSKLTMLADRAGLVMDRGSKITYNGVKIGRVADISETVRNGRPVAEFTLD
Mb0604|M.bovis_AF2122/97 KTELTMVASRAGLVMEAGSKVTYNGVEIGRVGSISEIERDGRPAAKLVLD
Rv0589|M.tuberculosis_H37Rv KTELTMVAFRAGLVMEAGSKVTYNGVEIGRVGSISEIERDGRPAAKLVLD
MAV_4551|M.avium_104 KTKLTMVATRAGLVMETGSKVTYNGVAIGRVADISEIERDGAPAAKLVLD
MSMEG_0134|M.smegmatis_MC2_155 KVQLTVLSGRAGLSMDPGSKVTFNGVPIGRLASIDVVEVDDNPEARLTLD
TH_0154|M.thermoresistible__bu RTQLTLLADRAGLVMDPGSKVTFNGVEIGRVAQIHADTSDDRTRANIVLD
Mflv_0711|M.gilvum_PYR-GCK RTQLTMISARSGLSLDSGSKVTFNGVEIGKVSEVEETTVGGEPRAKILLE
Mvan_0134|M.vanbaalenii_PYR-1 RTQLTMISARSGLSMDPGAKVTYNGVEIGRVGNVEETSVDGEPRAKIILD
MAB_4153c|M.abscessus_ATCC_199 TKPITVVSPRAGLVMEEGGKVKFHGLQIGKVTSISYAN----DQAKLELA
:*:.: *:** :: *.*:.::*: **:: : *.: *
MMAR_0412|M.marinum_M VYPRYLPLIPENVDAQIKATTVFGGKYVSLTTPEHPSNKKLSAHSVINAS
MUL_1062|M.ulcerans_Agy99 VYPRYLPLIPENVDAQIKATTVFGGKYVSLTTPEHPSNKKLSAHSVINAS
MLBr_02589|M.leprae_Br4923 VYPRYLSLIPANVNADIKATTVFGGKYVSLTTPEHPSQKRLTPQTVIDAR
Mb0604|M.bovis_AF2122/97 VNPRYISLIPVNVVADIEAATLFGNKYVALSAPKIPQQQRISSHDVIDVG
Rv0589|M.tuberculosis_H37Rv VNPRYISLIPVNVVADIEAATLFGNKYVALSAPKIPQQQRISSHDVIDVG
MAV_4551|M.avium_104 VDPRYVNLIPANVVATIEAATLFGNKYVSLSAPENPSRQRISPRDVIDAR
MSMEG_0134|M.smegmatis_MC2_155 VDPKYLDLIPENANVELRATTVFGNKYISFLSPKNPSAERLSASTPIRAQ
TH_0154|M.thermoresistible__bu VDPKYVDLIPSNADVEIKASTVFGNKYVAFASPPDPSLTPITSAATIDVT
Mflv_0711|M.gilvum_PYR-GCK VNPKYLKLIPRNVNADISATTVFGNKYVSFTSPENPQSERISASDVIDVT
Mvan_0134|M.vanbaalenii_PYR-1 VNPKYLDLIPSNVNADISATTVFGNKYVSFTTPEDPRPERISASDVIDVT
MAB_4153c|M.abscessus_ATCC_199 VNAKDLRLIPSNATVRIAGTTVFGAKSVEFLAPQKPSDSSLRPGATVTAE
* .: : *** *. . : .:*:** * : : :* * : . : .
MMAR_0412|M.marinum_M AVTTEINTLFETLTSISEKVDPVKLNLTLSAAAEALTGLGNRFGEAIANG
MUL_1062|M.ulcerans_Agy99 AVTIEINTLFETLTSISEKVDPVKLNLTLSAAAEALTGLGNRFGEAIANG
MLBr_02589|M.leprae_Br4923 SVTTEINTLFQTITLIAEKVDPIKLNLTLSAAAQSLAGLGERFGQSIVNG
Mb0604|M.bovis_AF2122/97 SVTTEFNTLFETITSIAEKVDPIELNATLSAVAQAPDGLGGKFGESIVNG
Rv0589|M.tuberculosis_H37Rv SVTTEFNTLFETITSIAEKVDPIELNATLSAVAQALDGLGGKFGESIVNG
MAV_4551|M.avium_104 SVTTEFNTLFETVTSIAEKVSPIELNATLSALAQALDGLGGKFGESIVNG
MSMEG_0134|M.smegmatis_MC2_155 GVTTEFNTLFETITAISEQVDPIKLNETLTAAAQALDGLGDKFGRSIVDG
TH_0154|M.thermoresistible__bu KVTTEFNTLFQTVVDIARQVDPIKLNQTLTATAQALDGLGDRFGESIIDG
Mflv_0711|M.gilvum_PYR-GCK SVTTEFNTLFETVVEIAEQVDPIKLNQTLTATAQALDGLGDRFGESIING
Mvan_0134|M.vanbaalenii_PYR-1 SVTTEFNTLFETVVSLADQVDPIKLNQTLTATAQALDGLGDRFGESIING
MAB_4153c|M.abscessus_ATCC_199 AVALEVNTVFENLTRLLGKIDPVNLNATLTAVGEGLRGNGDNFGQGLVEL
*: *.**:*:.:. : ::.*::** **:* .:. * * .**..: :
MMAR_0412|M.marinum_M NEILDDINPQMPQARRDIQQLAALGDVYADAAPDLFDFLNNSVPTARTIN
MUL_1062|M.ulcerans_Agy99 NEILDDINPQMPQARRDIQQLAALGDVYADAAPDLFDFLNNSVPTARTIN
MLBr_02589|M.leprae_Br4923 NSVLDDVNSQLPQARHDIQQLASLGDTYANSASDFFDFLNNSIVTSRTIN
Mb0604|M.bovis_AF2122/97 NQILAQLNPRLPQLGYDVRRLADLGEVYVDASPDLWSFLQNALTTARTLT
Rv0589|M.tuberculosis_H37Rv NQILAQLNPRLPQLGYDVRRLADLGEVYVDASPDLWSFLQNALTTARTLT
MAV_4551|M.avium_104 NQILAQLNPRMPQIRYDVRRLADLAVAYTKAAPDLLDFLSNAVTTARTLT
MSMEG_0134|M.smegmatis_MC2_155 NAILADVNPRMPQIRRDITGLANLGEVYADASPDLFDGLDNAVTTARTLN
TH_0154|M.thermoresistible__bu NAILDDLNPQMPQIRYDVQRLADLNAVYAAAAPDLFDGLENAVTTAQTLY
Mflv_0711|M.gilvum_PYR-GCK NQILAEINPQMPQIRRDNQLLADFGEVYSNAAPDLFDGLQNAVTTARTLN
Mvan_0134|M.vanbaalenii_PYR-1 NQILSEINPQMPQIRRDNQLLADLGDVYADAAPDLFDGLQNAVTTARTLN
MAB_4153c|M.abscessus_ATCC_199 DQYLDQLNPKLPALQQDFRQAGQVGQIYGDAAPDIVRILDNLPALSRTVV
: * ::*.::* * . . * ::.*: *.* ::*:
MMAR_0412|M.marinum_M AQQHDLDAALLAAVGFGNTGADVFERGGPYLARGAADLVPSAQLLDTYSP
MUL_1062|M.ulcerans_Agy99 AQQHDLDAALLAAVGFGNTGADVFERGGPYLARGAADLVPSAQLLDTYSP
MLBr_02589|M.leprae_Br4923 QQQKDLDQVLLAAVGFGNTGADIFSRSGPYLARGAADLVPTAQLLDTYSP
Mb0604|M.bovis_AF2122/97 SQQRDLDAALLAATGAGNTGEDVFARGGPYLARAAADLVPTATLLDTYSP
Rv0589|M.tuberculosis_H37Rv SQQRDLDAALLAATGAGNTGEDVFARGGPYLARAAADLVPTATLLDTYSP
MAV_4551|M.avium_104 RQQGDLDAALLAAVGVGHEGEDIFARGGPYLARGAADLVPTAELLDTYSP
MSMEG_0134|M.smegmatis_MC2_155 EQRGNLDQALVAAVGFGNTGGDIFERGGPYLVRGAQDLLPTSALLDEYSP
TH_0154|M.thermoresistible__bu QERGNLDQTLLASIGFGNTAGEVFERGGPYLVRGSEDLVTVTEILDYHSP
Mflv_0711|M.gilvum_PYR-GCK EQQGDVDQALMAAVGFGNTGGDIFERGGPYLVRGARDLVPTSALLDKYSP
Mvan_0134|M.vanbaalenii_PYR-1 EQQGDVDQALMAAVGFGNTGGDVFERGGPYLARGAEDLVPTTALLDKYSP
MAB_4153c|M.abscessus_ATCC_199 DQQQDLKATLLAAIGLGNNGYETLAPAEKDLTAALQRLRAPATLLGRYSP
:: ::. .*:*: * *: . : : . *. . * . : :*. :**
MMAR_0412|M.marinum_M ELFCTLRNYHDVEPQAYKFLGGNG-YSLNSHTTPLSALGMMLNPASLVPL
MUL_1062|M.ulcerans_Agy99 ELFCTLRNYHDVEPQAYKFLGGNG-YSLNSHTTPLSALGMMLNPASLVPL
MLBr_02589|M.leprae_Br4923 AIFCTLRNYHDIEPKVTSFLGGNG-YSINDHVELLSGLGFLLNPTSAISI
Mb0604|M.bovis_AF2122/97 ELFCMIRNFHDAAPKVADAVGGNG-YSLAAAGTILGAP------------
Rv0589|M.tuberculosis_H37Rv ELFCMIRNFHDAAPKVADAVGGNG-YSLAAAGTILGAP------------
MAV_4551|M.avium_104 ELFCMIRNFHDAAPEVAKAAGGNG-YSLAAAGSIVGAP------------
MSMEG_0134|M.smegmatis_MC2_155 ALFCTIRNYHDAAPKLAGALGGNG-YSLLTNSLVVGV-------------
TH_0154|M.thermoresistible__bu ALFCMIRNYHDVAPKVAASLGGNG-YSLRQHAFLVGP-------------
Mflv_0711|M.gilvum_PYR-GCK SFFCMMRNYHDVEPKIADALGGNG-YSLKTHSQVLGLG------------
Mvan_0134|M.vanbaalenii_PYR-1 SFFCMIRNYHDVEPKIADALGGNG-YSLKTHSEVLGLG------------
MAB_4153c|M.abscessus_ATCC_199 EIPCTLQAISKALVTFTPLIGGVRPGLFLTNSFPLGAP------------
: * :: . ** : :.
MMAR_0412|M.marinum_M L---ISQVGGLA----AGLVGGAPNPYIYPENLPRVNAHGGP--GGAPGC
MUL_1062|M.ulcerans_Agy99 L---ISQVGGLA----AGLVGGAPNPYIYPENLPRVNAHGGP--GGAPGC
MLBr_02589|M.leprae_Br4923 IGAGVLSQGLLAPLGLAGVVNGAANPYIYPENLPRVNARGGP--GGAPGC
Mb0604|M.bovis_AF2122/97 ------------------------NPYVYPDNLPRVNAHGGP--GGRPGC
Rv0589|M.tuberculosis_H37Rv ------------------------NPYVYPDNLPRVNAHGGP--GGRPGC
MAV_4551|M.avium_104 ------------------------NPYVYPDNLPRVNAHGGP--GGRPGC
MSMEG_0134|M.smegmatis_MC2_155 -----------------------GNPYVYPDNLPRVNAKGGP--EGRPGC
TH_0154|M.thermoresistible__bu -----------------------GNPYVYPDNLPRVNATGGP--GGRPGC
Mflv_0711|M.gilvum_PYR-GCK ----------------------AGNAYVYPDNLPRVNAKGGP--GGRPGC
Mvan_0134|M.vanbaalenii_PYR-1 ----------------------AGNAYVYPDNLPRVNAKGGP--GGRPGC
MAB_4153c|M.abscessus_ATCC_199 -------------------------VYTYPDSLPIVNASGGPNCRGLPNI
* **:.** *** *** * *.
MMAR_0412|M.marinum_M WQSITHDFWP-APELVMDTG-NSLAPYNHLETGSPFVLEYVWGRQVGDYT
MUL_1062|M.ulcerans_Agy99 WQSITHDFWP-APELVMDTG-NSLAPYNHLETGSPFVLEYVWGRQVGDYT
MLBr_02589|M.leprae_Br4923 WQHITRDLWP-TPELVMDTG-NSIAPYNHLDSGSPYAIEYVWGRQVGDNT
Mb0604|M.bovis_AF2122/97 WQTITRELWP-APYLVMDTG-ASLAPYNHVELGQPMFTEYVWGRQYGENT
Rv0589|M.tuberculosis_H37Rv WQTITRELWP-APYLVMDTG-ASLAPYNHVELGQPMFTEYVWGRQYGENT
MAV_4551|M.avium_104 WQKITRDLWP-APYLVMDTG-ASLAPYNHLEIGQPLATEFVWGRQYGENT
MSMEG_0134|M.smegmatis_MC2_155 WQPITRDLWP-FPYLVMDTG-ASIAPYNHFELGQPMFAEYVWGRQVGENT
TH_0154|M.thermoresistible__bu WQPITRDLWP-APYLVMDTG-ASLAPYNHIELGKPFAIEYVWGRQIGEYT
Mflv_0711|M.gilvum_PYR-GCK WQPITRDLWP-APFLVLDTG-ASVAPYNHMELGQPLFTEYVWGRQVGEHT
Mvan_0134|M.vanbaalenii_PYR-1 WQPITRDLWP-APFLVLDTG-ASIAPYNHMELGQPLLTEYVWGRQVGEHT
MAB_4153c|M.abscessus_ATCC_199 PTKMQNGSWFRSPFLVTDNAYIPYKPLEELQVNAPDTLQYLYNGAFAKRS
: . * * ** *.. . * :..: . * ::::. .. :
MMAR_0412|M.marinum_M INP
MUL_1062|M.ulcerans_Agy99 INP
MLBr_02589|M.leprae_Br4923 INP
Mb0604|M.bovis_AF2122/97 INP
Rv0589|M.tuberculosis_H37Rv INP
MAV_4551|M.avium_104 INP
MSMEG_0134|M.smegmatis_MC2_155 INP
TH_0154|M.thermoresistible__bu INP
Mflv_0711|M.gilvum_PYR-GCK INP
Mvan_0134|M.vanbaalenii_PYR-1 INP
MAB_4153c|M.abscessus_ATCC_199 EF-