For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NYDVPDELSVSTDGPVRIVTINRETDLNSVNEKLHWALANVWRQLAADKDAKVVVLTGAGRAFSAGGDLG WITTFLDDPVARDESIREGAQIIEEMLRFPLPVIAAVNGPAVGLGCSVALLCDILLISEDAYLADPHVSV GLVAGDGGAALWPLLTPIMRSREFLYTGDRIDAGKAVELGLASRSVAPGQLRSEAMTLAHRLAKQPVEAL QGTKRVVNMYLSQVLSGPLQAGFAAEQVSMRSDDHRARLLAFKKKAHK*
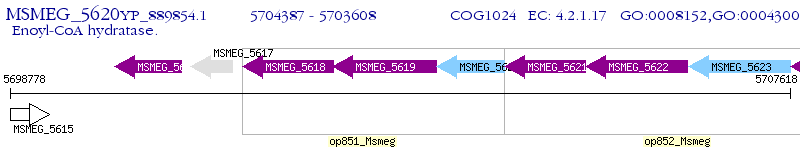
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5620 | - | - | 100% (259) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_4873 | - | 1e-60 | 46.72% (244) | enoyl-CoA hydratase/isomerase |
| M. smegmatis MC2 155 | MSMEG_3390 | - | 3e-30 | 36.06% (208) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2698 | echA15 | 7e-31 | 32.75% (229) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2395 | - | 7e-58 | 46.09% (243) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv2679 | echA15 | 7e-31 | 32.75% (229) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 7e-23 | 27.80% (259) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_0836c | - | 9e-25 | 30.43% (253) | putative enoyl-CoA hydratase/isomerase |
| M. marinum M | MMAR_4790 | - | 6e-61 | 47.56% (246) | enoyl-CoA hydratase |
| M. avium 104 | MAV_0867 | - | 3e-61 | 47.97% (246) | enoyl-CoA hydratase/isomerase family protein |
| M. thermoresistible (build 8) | TH_3045 | - | 1e-109 | 74.61% (256) | PUTATIVE Enoyl-CoA hydratase/isomerase |
| M. ulcerans Agy99 | MUL_0359 | - | 8e-62 | 47.97% (246) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4253 | - | 8e-58 | 45.27% (243) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5620|M.smegmatis_MC2_155 --MNYDVPDELSVSTDGP-----VRIVTINRETDLNSVNEKLHWALANVW
TH_3045|M.thermoresistible__bu --VKYDLPDVMTVRADGP-----IRTVTLDRAADLNSVDADLHWALANVW
MMAR_4790|M.marinum_M ---MYDMPTEIDVRAEGA-----LRIITLNRPDALNAVNDNLHVGLAGLW
MUL_0359|M.ulcerans_Agy99 ---MYDMPTEIDVRAEGA-----LRIITLNRPDALNAVNDNLHVGLAGLW
MAV_0867|M.avium_104 ---MYDMPTEIDVRADGA-----LRIITLNRPDSLNSVNDDLHVGLARLW
Mflv_2395|M.gilvum_PYR-GCK ---MYGMPDEIDVTADGD-----LRIITLNRPDDLNAVNDNLHVGLAKIW
Mvan_4253|M.vanbaalenii_PYR-1 ---MYGMPEEIDVRADGA-----LRIITLNRPDDLNAVNDNLHVGLAKIW
MLBr_02402|M.leprae_Br4923 -----MKYDTILVDGYQR-----VGIITLNRPQALNALNSQMMNEITNAA
MAB_0836c|M.abscessus_ATCC_199 ----MSAARELLVERDGP-----VVILTMNRPHRRNALSTNMVSQFAAAW
Mb2698|M.bovis_AF2122/97 MPVTYDDFPSLRCEIHDQPGHEGVLELVLDSP-GLNSVGPHMHRDLADIW
Rv2679|M.tuberculosis_H37Rv MPVTYDDFPSLRCEIHDQPGHEGVLELVLDSP-GLNSVGPHMHRDLADIW
: : :.:: *::. .: ::
MSMEG_5620|M.smegmatis_MC2_155 RQLAADKDAKVVVLTGAGRAFSAGGDLGWITTFLDDPVARDESIREGAQI
TH_3045|M.thermoresistible__bu RQLAADTDARVVILTGAGRAFSAGGDLDWITSFLTDQVARDESIREGAQI
MMAR_4790|M.marinum_M RRLSEDRSARAAVLTGAGRAFSAGGDFAYLAELSQDADLRAKTIAHGRDI
MUL_0359|M.ulcerans_Agy99 RRLSEDRSARAAVLTGAGRAFSAGGDFAYLAELAQDADLRAKTIAHGRDI
MAV_0867|M.avium_104 QRLTDDPTARAAVITGAGRAFSAGGDFGYLKELSADADLRAKTIRDGREI
Mflv_2395|M.gilvum_PYR-GCK EELNEDPGARAAVITGAGRAFSAGGDFHYLDELRRDEALRQKTIKHGRDL
Mvan_4253|M.vanbaalenii_PYR-1 EELNEDAGARAAVITGAGRAFSAGGDFNYLDELRNDEALRQKTIKHGRDL
MLBr_02402|M.leprae_Br4923 KELDIDPDVGAILITGSPKVFAAGADIKE--MASLTFTDAFDADFFSAWG
MAB_0836c|M.abscessus_ATCC_199 DEIDHDDGIRAAILTGAGSAYCVGGDLSDGWMVRDGSAPPLDPATIGKGL
Mb2698|M.bovis_AF2122/97 PVIDRDPAVRVVLVRGEGKAFSSGGSFDLIAETIGDYQGRLRIMREARDL
Rv2679|M.tuberculosis_H37Rv PVIDRDPAVRVVLVRGEGKAFSSGGSFDLIAETIGDYQGRLRIMREARDL
: * . :: * .:. *..: .
MSMEG_5620|M.smegmatis_MC2_155 IEEMLRFPLPVIAAVNGPAVGLGCSVALLCDILLISEDAYLADPHVSVGL
TH_3045|M.thermoresistible__bu IEEMLRFPLPVIAAVNGPAVGLGCSVAVLCDIVLIAENAYLADPHVAVGL
MMAR_4790|M.marinum_M VLGMARCRVPVVAAVNGPAVGLGCSLVALSDIVYIAPDAYLADPHVQVGL
MUL_0359|M.ulcerans_Agy99 VLGMARCRVPVVAAVNGPAVGLGCSLVALSDIVYIAPDAYLADPHVQVGL
MAV_0867|M.avium_104 VLGMARCRIPVVAAVNGPAVGLGCSLVALSDIVYIAENAYLADPHVQVGL
Mflv_2395|M.gilvum_PYR-GCK VIGMVRCRIPVIAAVNGPAVGLGCSLAALSDVVYIAENAFFADPHVSIGL
Mvan_4253|M.vanbaalenii_PYR-1 VIGMVRCRIPVVAAVNGPAVGLGCSLAALSDVVYIAENAFFADPHVSIGL
MLBr_02402|M.leprae_Br4923 KLAAVRT--PMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGV
MAB_0836c|M.abscessus_ATCC_199 LLSHTLTK-PLIAAVNGACLGGGCEMLQQTDIRVSDEHATFGLPEVQRGL
Mb2698|M.bovis_AF2122/97 VLNLVNFDKPVVSAIRGPAVGAGLVVALLADISVAGRAAKIIDGHTKLGV
Rv2679|M.tuberculosis_H37Rv VLNLVNFDKPVVSAIRGPAVGAGLVVALLADISVAGRAAKIIDGHTKLGV
*:::*: * .:* * : *: * : . *:
MSMEG_5620|M.smegmatis_MC2_155 VAGDGGAALWPLLTPIMRSREFLYTGDRIDAGKAVELGLASRSVAPGQLR
TH_3045|M.thermoresistible__bu VAGDGGAAMWPLLTLVLRSREYLFTGDRIPAATAVELGLASRTVPADDLL
MMAR_4790|M.marinum_M VAADGGPLTWPLHISLMLAKEYALTGQRIAAQRAVELGLVN--HVAADPV
MUL_0359|M.ulcerans_Agy99 VAADGGPLTWPLHISLMLAKEYALTGQRIAAQRAVELGLAN--HVAADPV
MAV_0867|M.avium_104 VAADGGPLTWPLHISLLLAKEYALTGTRISAQRAVELGLAN--HVADDPV
Mflv_2395|M.gilvum_PYR-GCK VAADGGPLVWGSQISLLQAKEFALTGVRIKAQKAVELGLAN--HVVDDPV
Mvan_4253|M.vanbaalenii_PYR-1 VAADGGPLVWGSQISLLQAKEFALTGVRIKAQRAVELGLAN--HVVDDPV
MLBr_02402|M.leprae_Br4923 LPGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLL
MAB_0836c|M.abscessus_ATCC_199 VPGAGSMVRLKRQIPYTKAMEMILTGEPLTAFEAYHFGLVGHVVPAGTAL
Mb2698|M.bovis_AF2122/97 AAGDHAAICWPLLVGMAKAKYYLLTCEPLSGEEAERIGLVSICVDDDDVL
Rv2679|M.tuberculosis_H37Rv AAGDHAAICWPLLVGMAKAKYYLLTCEPLSGEEAERIGLVSICVDDDDVL
.. . : * : . * . **..
MSMEG_5620|M.smegmatis_MC2_155 SEAMTLAHRLAKQPVEALQGTKRVVNMYLSQVLSGPLQAGFAAEQVSMRS
TH_3045|M.thermoresistible__bu PEAIRLAERLAAQPAEALRGTKRIVNMYLSQALAGPLQAGFAAEVATMRT
MMAR_4790|M.marinum_M AEAVACAQRIMKLPQQAVESTKRVLNIHLERAVLASLDYALSAEHQSFTT
MUL_0359|M.ulcerans_Agy99 AEAVACAQRIVKLPQQAVESTKRVLNIHLERAVLASLDYALSAEHQSFTT
MAV_0867|M.avium_104 AEAIACAQKILELPQQAVESTKRVLNIHLERAVLASLDYALSAESQSFVT
Mflv_2395|M.gilvum_PYR-GCK GEAIACAKKIMALPQQAVEATKRLMNIQLEKSVMASLDYANLAEYVSFGT
Mvan_4253|M.vanbaalenii_PYR-1 AEAIACAKKLIELPQQAVEATKRLMNIQLEKSVMASLDYANLAEYVSFGT
MLBr_02402|M.leprae_Br4923 PEAKAVATTISQMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFVT
MAB_0836c|M.abscessus_ATCC_199 DKARSLADRIVRNGPLAVRNAKEAIVRSGWLAEEDARAIEARLTRPVITS
Mb2698|M.bovis_AF2122/97 PTATRLAERLAAGAQNAIRWTKRSLN-HWYRMFGPAFETSLGLEFIGFGG
Rv2679|M.tuberculosis_H37Rv PTATRLAERLAAGAQNAIRWTKRSLN-HWYRMFGPAFETSLGLEFIGFGG
* * : * . :*. : :
MSMEG_5620|M.smegmatis_MC2_155 DDHRARLLAFKKKAHK------------
TH_3045|M.thermoresistible__bu AEHRDRLLALRERTR-------------
MMAR_4790|M.marinum_M EDFRSIVTRLTAPKN-------------
MUL_0359|M.ulcerans_Agy99 EDFRSIVTRLTAPKN-------------
MAV_0867|M.avium_104 EDFRSIVTKL-ADKN-------------
Mflv_2395|M.gilvum_PYR-GCK ADFNRIVDGLIAKN--------------
Mvan_4253|M.vanbaalenii_PYR-1 ADFNKIVDGLIAKK--------------
MLBr_02402|M.leprae_Br4923 DDQSEGMAAFIEKRAPQFTHR-------
MAB_0836c|M.abscessus_ATCC_199 ADAREGLAAFKEKREARFTGR-------
Mb2698|M.bovis_AF2122/97 PDVREGLAAHREKRPARFGADPDPGAGS
Rv2679|M.tuberculosis_H37Rv PDVREGLAAHREKRPARFGADPDPGAGS
: :