For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVIVTGGSRGIGRQIVERLVRDGHDVVFTHSDSEADAAEVERTCGAGARGVRLDVTDADAPGRLFDTAE ATGTVTALVNNAGVTGPLGPLTALDDDALRRTVEVNLVAAARLCREAAQRWQSRPAERRDIVNVSSVAAR TGSPGEYVVYAATKAALDTLTLGLAKELGPSGVRVNSVRAGTTDTTIHARAGDPDRPRRVARHVPLGRVA EPGEIAAAVAWLLSADAAYVSGSVLDVTGGL
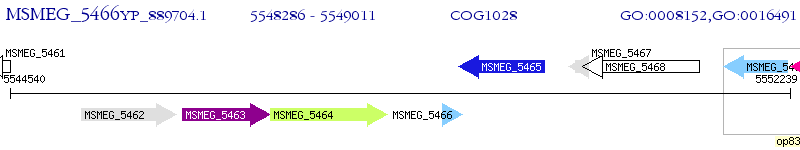
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5466 | - | - | 100% (241) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_2598 | - | 1e-29 | 35.54% (242) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2029 | - | 1e-28 | 36.89% (244) | 3-ketoacyl-ACP/CoA redutase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1519 | fabG1 | 1e-30 | 35.68% (241) | 3-oxoacyl- |
| M. gilvum PYR-GCK | Mflv_3658 | - | 1e-29 | 36.51% (241) | 3-oxoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv1483 | fabG1 | 2e-30 | 35.68% (241) | 3-oxoacyl- |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 4e-31 | 36.10% (241) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_2723c | - | 3e-34 | 35.68% (241) | 3-oxoacyl- |
| M. marinum M | MMAR_2289 | fabG1 | 3e-29 | 35.27% (241) | 3-oxoacyl- |
| M. avium 104 | MAV_3714 | - | 4e-30 | 35.34% (249) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_2925 | - | 8e-27 | 36.33% (245) | PUTATIVE putative short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1491 | fabG1 | 2e-29 | 35.27% (241) | 3-oxoacyl- |
| M. vanbaalenii PYR-1 | Mvan_2752 | - | 2e-28 | 35.27% (241) | 3-oxoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2289|M.marinum_M MTDTATEQNTESAADGGKPAFVSRSVLVTGGNRGIGLAIAQRLAADGHRV
MUL_1491|M.ulcerans_Agy99 MTDTATEQNTESAADGGKPAFVSRSVLVTGGNRGIGLAIAQRLATDGHRV
Mb1519|M.bovis_AF2122/97 MTATATEG--------AKPPFVSRSVLVTGGNRGIGLAIAQRLAADGHKV
Rv1483|M.tuberculosis_H37Rv MTATATEG--------AKPPFVSRSVLVTGGNRGIGLAIAQRLAADGHKV
Mflv_3658|M.gilvum_PYR-GCK MTSTEN---TAGESVAGRPAFVSRSVLVTGGNRGIGLAIAQRLAADGHKV
Mvan_2752|M.vanbaalenii_PYR-1 MTLTENSAGTAGETGGGRPAFVSRSVLVTGGNRGIGLAIAQRLAADGHKV
MLBr_01807|M.leprae_Br4923 MTDAAAKV--SAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRV
MAB_2723c|M.abscessus_ATCC_199 MTNA---------------EFVPRSVLVTGGNRGIGLAVAQRLAADGHKV
MAV_3714|M.avium_104 -------------MMDLTQRLAGRVAVITGAGGGIGLAAARRMHAEGATI
TH_2925|M.thermoresistible__bu -----------------MSSEVVPVAVVTGGARGIGEAIAAALIDAGYRT
MSMEG_5466|M.smegmatis_MC2_155 -----------------------MTVIVTGGSRGIGRQIVERLVRDGHDV
::**. *** . : *
MMAR_2289|M.marinum_M AVTHR----GSGAPEGLFG-------VECDVTDNDAVDRAFKEVEEHQGP
MUL_1491|M.ulcerans_Agy99 AVTHR----GSGAPEGLFG-------VECDVTDNDAVDRAFKEVEEHQGP
Mb1519|M.bovis_AF2122/97 AVTHR----GSGAPKGLFG-------VECDVTDSDAVDRAFTAVEEHQGP
Rv1483|M.tuberculosis_H37Rv AVTHR----GSGAPKGLFG-------VECDVTDSDAVDRAFTAVEEHQGP
Mflv_3658|M.gilvum_PYR-GCK AVTHR----GSGAPDGLFG-------VVCDVTDNDAVDRAFKEVEEHQGP
Mvan_2752|M.vanbaalenii_PYR-1 AVTHR----GSGAPEGLFG-------VECDVTDNEAVDRAFAEVEEHQGP
MLBr_01807|M.leprae_Br4923 AVTHR----ASVVPDWFFG-------VECDVTDNDAIGAAFKAVEEHQGP
MAB_2723c|M.abscessus_ATCC_199 AVTHR----GSGAPEGLLG-------VQCDITDNDQIDAAFKEVEEKQGP
MAV_3714|M.avium_104 VVADIDADAGAAAADELSG-----LFVPTDVADEDAVNALFDTAVRRYGR
TH_2925|M.thermoresistible__bu AVIDVNEKAITRAADSDAARQGKLIPLQLSITDRPAVERAMGELADAHGP
MSMEG_5466|M.smegmatis_MC2_155 VFTHSDSEADAAEVERTCG--AGARGVRLDVTDADAPGRLFDT-AEATGT
.. . : . . : .::* : *
MMAR_2289|M.marinum_M VEVLVSNAGLSADAF--LIRMTEERFEKVIDANLTGAFRVAQRASR--SM
MUL_1491|M.ulcerans_Agy99 VEVLVSNAGLSADAF--LIRMTEERFEKVIDANLTGAFRVAQRASR--SM
Mb1519|M.bovis_AF2122/97 VEVLVSNAGLSADAF--LMRMTEEKFEKVINANLTGAFRVAQRASR--SM
Rv1483|M.tuberculosis_H37Rv VEVLVSNAGLSADAF--LMRMTEEKFEKVINANLTGAFRVAQRASR--SM
Mflv_3658|M.gilvum_PYR-GCK VEVLVSNAGISKDAF--LMRMTEDRFTDVIDANLTGAFRVAQRASR--SM
Mvan_2752|M.vanbaalenii_PYR-1 VEVLVSNAGISKDAF--LMRMTEERFEEVINANLTGAFRVAQRASR--SM
MLBr_01807|M.leprae_Br4923 VEVLVANAGITSDML--IMRMSEEQFQKVIDVNLTGVFRVAKLASR--NM
MAB_2723c|M.abscessus_ATCC_199 VEVLVANAGITADML--IMRMSEEQFLKVIDANLIGTFRLAKRASR--NM
MAV_3714|M.avium_104 IDIAFNNAGISPPDDDVIENTEPPAWQRVQDVNLKSVYLCCRAALR--HM
TH_2925|M.thermoresistible__bu IRALVNNAGMTLPG--SFLDQTDEDWDRIVDVNLKGTFVMSQVVAR--QM
MSMEG_5466|M.smegmatis_MC2_155 VTALVNNAGVTGPLG-PLTALDDDALRRTVEVNLVAAARLCREAAQRWQS
: . ***:: : :.** .. .: . :
MMAR_2289|M.marinum_M QRKKFGRLIFIGSVSGSWGIG-NQANYAASKAGVIGMARSIARELSKVNV
MUL_1491|M.ulcerans_Agy99 QRKKFGRLIFIGSVSGSWGIG-NQANYAASKAGVIGMARSIARELSKVNV
Mb1519|M.bovis_AF2122/97 QRNKFGRMIFIGSVSGSWGIG-NQANYAASKAGVIGMARSIARELSKANV
Rv1483|M.tuberculosis_H37Rv QRNKFGRMIFIGSVSGSWGIG-NQANYAASKAGVIGMARSIARELSKANV
Mflv_3658|M.gilvum_PYR-GCK QRKRFGRIIFIGSVSGMWGIG-NQANYAAAKAGLIGMARSISRELSKAGV
Mvan_2752|M.vanbaalenii_PYR-1 QRKRFGRIIFIGSVSGMWGIG-NQANYAAAKAGLIGMARSISRELSKANV
MLBr_01807|M.leprae_Br4923 IKQRFGRYIFMGSVVGLSGVP-GQANYAASKSGLIGMARSLARELGSRSI
MAB_2723c|M.abscessus_ATCC_199 IKKRFGRLIFMGSVAGQIGLP-GQANYSASKAGLVGMARALTRELGSRSI
MAV_3714|M.avium_104 VSARRGSIINTASFVAVMGSATSQISYTAAKGGVLALSRELGVQFARQGI
TH_2925|M.thermoresistible__bu VEERTGAIVN-ISSVSAHGVRTGPPAYAAAKAGVEGLSRLMAVQLGPSGI
MSMEG_5466|M.smegmatis_MC2_155 RPAERRDIVNVSSVAARTGSPGEYVVYAATKAALDTLTLGLAKELGPSGV
. : * . * *:*:*..: :: : ::. .:
MMAR_2289|M.marinum_M TANVVAPGYIDT---DMTRALDERIQEGALQFIPAKRVGTAAEVAGVVSF
MUL_1491|M.ulcerans_Agy99 TANVVAPGYIDT---DMTRALDERIQEGALQFIPAKRVGTAAEVAGVVSF
Mb1519|M.bovis_AF2122/97 TANVVAPGYIDT---DMTRALDERIQQGALQFIPAKRVGTPAEVAGVVSF
Rv1483|M.tuberculosis_H37Rv TANVVAPGYIDT---DMTRALDERIQQGALQFIPAKRVGTPAEVAGVVSF
Mflv_3658|M.gilvum_PYR-GCK TANVVAPGYIDT---EMTRALDERIQAGALEFIPAKRVGTADEVAGAVSF
Mvan_2752|M.vanbaalenii_PYR-1 TANVVAPGYIDT---EMTRALDERIQAGALEFIPAKRVGTADEVAGAVSF
MLBr_01807|M.leprae_Br4923 TANVVAPGFIDT---EMTRAMDSKYQEAALQAIPLGRIGAVEEIAGAVSF
MAB_2723c|M.abscessus_ATCC_199 TANVVAPGFIDT---DMTRELGEKHVETALTAIPMGRIGEPSEVAGVVSW
MAV_3714|M.avium_104 RVNALCPGPVNTPLLQELFAKDPQRAARRLVHVPVGRFAEPGEIAAAAAF
TH_2925|M.thermoresistible__bu RVNTVVVGTTATPWLLSRKTDEELATMR--ESTLLDRLGEPEDVAATVAF
MSMEG_5466|M.smegmatis_MC2_155 RVNSVRAGTTDT--TIHARAGDPDRPRRVARHVPLGRVAEPGEIAAAVAW
.* : * * *.. ::*...::
MMAR_2289|M.marinum_M LASEDASYISGAVIPVDGGMGMGH----
MUL_1491|M.ulcerans_Agy99 LASEDASYISGAVIPVDGGMGMGH----
Mb1519|M.bovis_AF2122/97 LASEDASYISGAVIPVDGGMGMGH----
Rv1483|M.tuberculosis_H37Rv LASEDASYISGAVIPVDGGMGMGH----
Mflv_3658|M.gilvum_PYR-GCK LASEDASYIAGAVIPVDGGMGMGH----
Mvan_2752|M.vanbaalenii_PYR-1 LASEDASYIAGAVIPVDGGMGMGH----
MLBr_01807|M.leprae_Br4923 LASEDAGYISGAVIPVDGGLGMGH----
MAB_2723c|M.abscessus_ATCC_199 LASPDAAYVSGAVIPVDGGLGMGH----
MAV_3714|M.avium_104 LASDDASFITASTFLVDGGISAAYVTPL
TH_2925|M.thermoresistible__bu LCSPAARHVTGQLVSVSGGQWMP-----
MSMEG_5466|M.smegmatis_MC2_155 LLSADAAYVSGSVLDVTGGL--------
* * * .::. . * **