For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGAMGRPRDPSKDVAVLTAVRELLVEEGYQGTTVLAVARRAGVGAPTIYRRWPTKEALVEDAAFGHPSP VPMPAPTGDLRADLREWVAMFLDWLAQPVTRAALPGLITAYHHDETLYERLVLRSETDVRAAMVDLLDGD RRRADAVFDFLVATTVVRAMTRGLTDRDAFCDRTADALFALARSDHL
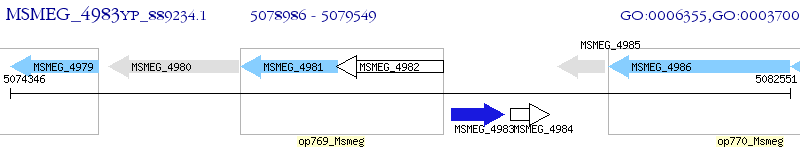
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4983 | - | - | 100% (187) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2043 | - | 5e-22 | 43.07% (137) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1500 | - | 2e-14 | 31.91% (188) | TetR family protein transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3192c | - | 1e-22 | 38.24% (170) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4496 | - | 7e-24 | 40.32% (186) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3167c | - | 1e-22 | 38.24% (170) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00316 | - | 6e-05 | 48.84% (43) | putative TetR/AcrR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0832 | - | 5e-32 | 41.97% (193) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0340 | - | 3e-23 | 38.42% (190) | TetR family transcriptional regulator |
| M. avium 104 | MAV_5165 | - | 5e-26 | 39.23% (181) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3283 | - | 2e-38 | 43.32% (187) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_4780 | - | 1e-22 | 37.89% (190) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1870 | - | 2e-23 | 39.78% (186) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3192c|M.bovis_AF2122/97 ----MKADLPSLDKAPGAGRPRDPRIDSAILSATAELLVQIGYSNLSLAA
Rv3167c|M.tuberculosis_H37Rv ----MKADLPSLDKAPGAGRPRDPRIDSAILSATAELLVQIGYSNLSLAA
Mflv_4496|M.gilvum_PYR-GCK ----MKTDSSAGGKATGAGRPRDPRIDAAILAATADLLVEIGYSNVTMAA
Mvan_1870|M.vanbaalenii_PYR-1 ----MNTDPSPGDKTPGAGRPRDPRIDAAILRATADLLVEIGYSNVTMAA
MMAR_0340|M.marinum_M ----MVAD---------FGRPRDPRIDGAVLRATVELLAETGYAGLLVSA
MUL_4780|M.ulcerans_Agy99 ----MVAD---------FGRPRDPRIDGAVLRATVELLAETGYAGLLVSA
MAV_5165|M.avium_104 ------MD---------FGRPRDPRIDAAVLRATVELLAETGYPGLLVSA
MSMEG_4983|M.smegmatis_MC2_155 ----MTG---------AMGRPRDPSKDVAVLTAVRELLVEEGYQGTTVLA
TH_3283|M.thermoresistible__bu ----MPGS-----GARSPGRPRDPALDDAIRAAARDVVVEHGYDATTLAL
MAB_0832|M.abscessus_ATCC_1997 ----MDKR--------QGGRPRDPDKDDAVRETVRRMLAADGYQRTTIPA
MLBr_00316|M.leprae_Br4923 MSVPRSGLSVQMAQPARPLRADAARNRARILGVAYEAFATEGLS-VPIDE
*. . : .. .. * :
Mb3192c|M.bovis_AF2122/97 VAERAGTTKSALYRRWSSKAELVHEAAF-PAAPTALQAAAGDIAADIRMM
Rv3167c|M.tuberculosis_H37Rv VAERAGTTKSALYRRWSSKAELVHEAAF-PAAPTALQAAAGDIAADIRMM
Mflv_4496|M.gilvum_PYR-GCK VAERAGTTKTALYRRWSSKAELVHEAAF-PVTPTAIQSPPGDIAADLRAM
Mvan_1870|M.vanbaalenii_PYR-1 VAERAGTTKTALYRRWSSKAELVHEAAF-PVTPTAIQTPPGDIAADLRAM
MMAR_0340|M.marinum_M IAKRAGTSKPAIYRRWPSKAHLVHEAVF-PIDAATAVPDTGSPVDDLREM
MUL_4780|M.ulcerans_Agy99 IAKRAGTSKPAIYRRWPSKAHLVHEAVF-PIDAATAVPDTGSPVDDLREM
MAV_5165|M.avium_104 IAQRAGTSKPAIYRRWPSKAHLVHEAVF-PVGADTAIPDTGSTADDLREM
MSMEG_4983|M.smegmatis_MC2_155 VARRAGVGAPTIYRRWPTKEALVEDAAFGHPSPVPMPAPTGDLRADLREW
TH_3283|M.thermoresistible__bu IADRAGVSVPTIYRRWQHKHELIEEAVL-HLDTFELPEPTGDLEADLGTW
MAB_0832|M.abscessus_ATCC_1997 VARAAGIGAPTIYRRWPTQAAMVESAIADRPWPEELAAPS-EFRYYLPVL
MLBr_00316|M.leprae_Br4923 IARRAGVGAGTVYRHFPTKEALCAAVIG---DRMPHLVDDGYVLLKSAGP
:* ** ::**:: : : .
Mb3192c|M.bovis_AF2122/97 IAATRDVFTTPVVRAALP--GLVADMTADAELNAR-VLARFADLFAA-VR
Rv3167c|M.tuberculosis_H37Rv IAATRDVFTTPVVRAALP--GLVADMTADAELNAR-VLARFADLFAA-VR
Mflv_4496|M.gilvum_PYR-GCK IEVTRDVFLSPVVRAALP--GLIADMSANADLTGR-IGARFTDLFAA-VR
Mvan_1870|M.vanbaalenii_PYR-1 IEAARDVFTSPVVRSALP--GLIADLSADADLAGR-VGARFTDLFAA-VR
MMAR_0340|M.marinum_M VRRTTAVLSTPAAKAALP--GLIGEMAADPTLHSA-LLVRFGEIFGHGLT
MUL_4780|M.ulcerans_Agy99 VRRTTAVLSTPAAKAAQP--GLIGEMAADPTLHSA-LLVRFGEIFGHGLT
MAV_5165|M.avium_104 VRRTMVFLTTPAAKAALP--GLIGEMAADPSLHSA-LLERFAGAIGGGLA
MSMEG_4983|M.smegmatis_MC2_155 VAMFLDWLAQPVTRAALP--GLITAYHHDETLYER-LVLRSETDVRAAMV
TH_3283|M.thermoresistible__bu VRLFLTVAMEPAARAAVP--GLMSAYSRNPDDYRR-LMTRGELPVRAAIA
MAB_0832|M.abscessus_ATCC_1997 VRAVVGYFADPATRAAMP--GLLVEYYREPSQYVV-LAERTEMPLREAFR
MLBr_00316|M.leprae_Br4923 GEALFTYLRSLVLHWGATDRGLVDALAGAGGIDILGVAPDAEDAFLTILS
. : . . **: : . .
Mb3192c|M.bovis_AF2122/97 MRLREAVDRGEAHPDVDPDRLIELIGGATMLR-MLLYPDDMLD---DAWV
Rv3167c|M.tuberculosis_H37Rv MRLREAVDRGEAHPDVDPDRLIELIGGATMLR-MLLYPDDMLD---DAWV
Mflv_4496|M.gilvum_PYR-GCK ARLQDAVERGEVRESVDPDRLVSLVGGATLLA-LLQTPDALAS---PSWV
Mvan_1870|M.vanbaalenii_PYR-1 NRLHDAVERGEVRAGVDPDQLVNLIGGATLLA-LLQAPDDLTA---PAWV
MMAR_0340|M.marinum_M TWLDNAAARGQVRADVTAPELAEAIAGIVLIG-LLTRPGELEPGVGDAWI
MUL_4780|M.ulcerans_Agy99 TWLDNAAARGQVRADVTAPELAEAIAGIVLIG-LLTRPGELEPGVGDAWI
MAV_5165|M.avium_104 DWLAAAAARGEARPDVTAAELAETIAGVTLVA-LLTRPTELD----DAWV
MSMEG_4983|M.smegmatis_MC2_155 DLLDGDRRR--------ADAVFDFLVATTVVR-AMTRGLTDRD----AFC
TH_3283|M.thermoresistible__bu AMLERAKAAGQAAPDCDPDAVFELIRGATFLR-ALTHSDEDAE----AFC
MAB_0832|M.abscessus_ATCC_1997 RAHAGAVGSGERADQPGADALFDTIVGMAVYHGVLRGTADDALVEQILDV
MLBr_00316|M.leprae_Br4923 DLLYAAQYAGTARTDVGVREVKSILVGCQAMEAYNS-----------ALA
: . : .
Mb3192c|M.bovis_AF2122/97 DQTTAIVVRGVHRAAPGGSVV---
Rv3167c|M.tuberculosis_H37Rv DQTTAIVVRGVHRAAPGGSVV---
Mflv_4496|M.gilvum_PYR-GCK DSTTDILLRGVVAGA---------
Mvan_1870|M.vanbaalenii_PYR-1 DRTTDILLHGVMA-----------
MMAR_0340|M.marinum_M DRTTTLLLKGIRKGTPQ-------
MUL_4780|M.ulcerans_Agy99 DRTTTLLLKGIRKGTPQ-------
MAV_5165|M.avium_104 DRTTRLLLKGISA-----------
MSMEG_4983|M.smegmatis_MC2_155 DRTADALFALARSDHL--------
TH_3283|M.thermoresistible__bu TRITEAVLRAVRS-----------
MAB_0832|M.abscessus_ATCC_1997 VDAASSVGRGLHLDETNSDKERKR
MLBr_00316|M.leprae_Br4923 ERVTDVVVDGLRAAR---------
: :