For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YVSGGMNFELTEDQQLIRSSVAELAAKFDDHYWMAKDQAHEFPQEFYDAIAGGGWLGMTIPEQYGGHGLG ITEATILAEEVARSGGGMNAASAIHMSIFGMQPVVVFGSDEMKAETLPRIVDGDLHVCFGVTEPGAGLDT SRITTFARRDGDHPDADYVVNGRKVWISKALESEKILLLTRTESREDVEKRGGKPTEGLSLFLTDLDRDH VDIRPINKMGRNAVSSNELFIDDLRVPVEHRIGEEGKGFGYILHGLNPERMLIAAEALGIGRVALDRAVK YANDRVVFDRPIGMNQGIQFPLADSLARLDAAELVLRKATWLYDNGKPCGREANMAKYLCADAGFAAADR ALQTHGGMGYSEEYNISRFFRESRLMKIAPVSQEMILNFLGANVLGLPKSY*
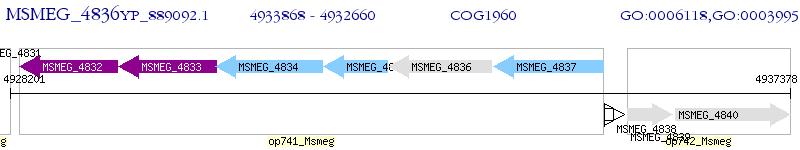
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4836 | - | - | 100% (402) | FadE12_1 |
| M. smegmatis MC2 155 | MSMEG_5494 | - | 6e-70 | 38.50% (400) | acyl-CoA dehydrogenase fadE12 |
| M. smegmatis MC2 155 | MSMEG_0338 | - | 1e-68 | 38.04% (397) | acyl-CoA dehydrogenase fadE12 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0997c | fadE12 | 3e-71 | 37.50% (400) | acyl-CoA dehydrogenase FADE12 |
| M. gilvum PYR-GCK | Mflv_2442 | - | 0.0 | 85.14% (397) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv0972c | fadE12 | 6e-71 | 37.50% (400) | acyl-CoA dehydrogenase FADE12 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 8e-46 | 30.83% (373) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2065c | - | 2e-72 | 39.55% (397) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_4741 | fadE12_1 | 0.0 | 80.56% (396) | acyl-CoA dehydrogenase FadE12_1 |
| M. avium 104 | MAV_1088 | - | 3e-71 | 37.50% (400) | acyl-CoA dehydrogenase fadE12 |
| M. thermoresistible (build 8) | TH_4562 | - | 0.0 | 89.14% (396) | PUTATIVE FadE12_1 |
| M. ulcerans Agy99 | MUL_4707 | fadE12 | 4e-72 | 37.53% (405) | acyl-CoA dehydrogenase FadE12 |
| M. vanbaalenii PYR-1 | Mvan_4210 | - | 0.0 | 84.92% (398) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0997c|M.bovis_AF2122/97 -------MTDTSFIESEERQALRKAVASWVANYGHEYYLDKARKHEHTSE
Rv0972c|M.tuberculosis_H37Rv -------MTDTSFIESEERQALRKAVASWVANYGHEYYLDKARKHEHTSE
MUL_4707|M.ulcerans_Agy99 ------MMTGTSFIENEDRRALRKAVSEWASNYGHEYYLKKARAHEHTTE
MAV_1088|M.avium_104 -------MTDSSFIENEERQALRKAVAEWASNYGSEYYLRKARAHEHTNE
MAB_2065c|M.abscessus_ATCC_199 ----------MELHETEERQDLRKAVAEIAKDFGHEYYLEKSLAGAKSTE
MSMEG_4836|M.smegmatis_MC2_155 ----MYVSGGMNFELTEDQQLIRSSVAELAAKFDDHYWMAKDQAHEFPQE
TH_4562|M.thermoresistible__bu ----------MHFELTEDQELIRASVRELAAKFDDTYWMEKDLNHEFPQE
Mflv_2442|M.gilvum_PYR-GCK ------MPG-MSFELTEDQELIRKSVAELCSKFDDHYWMEKDQNHEFPQE
Mvan_4210|M.vanbaalenii_PYR-1 ---MTSMPGGMSFELTEDQELIRKSVRELASRFDDHYWMEKDQAHEFPQE
MMAR_4741|M.marinum_M ----------MDFELTEDQELIRRSVAELASRFDDHYWMEKDQAHEFPAE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
: *::. :* :: : . *
Mb0997c|M.bovis_AF2122/97 LWAEAGKLGFLGVNLPEEYGGGGAGMYELSLVMEEMAAAGSAL-LLMVVS
Rv0972c|M.tuberculosis_H37Rv LWAEAGKLGFLGVNLPEEYGGGGAGMYELSLVMEEMAAAGSAL-LLMVVS
MUL_4707|M.ulcerans_Agy99 LWSEAGKLGFIGVNLPEEYGGGGAGMYELSMVMEEMAAAGSAL-LLMVVS
MAV_1088|M.avium_104 LWSEAGKLGFLGVNLPEEYGGGGAGMYELSLVMEEMAAAGSAL-LLMVVS
MAB_2065c|M.abscessus_ATCC_199 LWQAVGKQGFLGVNLSEQYGGGGGGIYDMQIVGEELAAAGCPL-LMTVVS
MSMEG_4836|M.smegmatis_MC2_155 FYDAIAGGGWLGMTIPEQYGGHGLGITEATILAEEVARSGGGMNAASAIH
TH_4562|M.thermoresistible__bu FYDAIAGGGWLGMTIPEEYGGHGLGITEATILAEEVARSGGGMNAASSIH
Mflv_2442|M.gilvum_PYR-GCK FYDAISKGGWLGMTIPEEYGGHGLGITEATLLLEEVSRSGAAMNGASAIH
Mvan_4210|M.vanbaalenii_PYR-1 FYDAIANGGWLGMTIPEEYGGHGLGITEATLLLEEVSRSGAAMNGASAIH
MMAR_4741|M.marinum_M FYRAIADGGWLGMTIPTEYGGHGLGITEATILLEEVAKSGGAMNAASSIH
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
*: .: :. :*** * . :: **:: . :
Mb0997c|M.bovis_AF2122/97 PAING-TIIAKFGTDDQKKRWLPGIADGSLTMAFAITEPDAGSNSHKITT
Rv0972c|M.tuberculosis_H37Rv PAING-TIIAKFGTDDQKKRWLPGIADGSLTMAFAITEPDAGSNSHKITT
MUL_4707|M.ulcerans_Agy99 PAING-TIISKFGTDEQKQRWLPGIADGTLTMAFAITEPEAGSNSHKITT
MAV_1088|M.avium_104 PAING-TIISKFGTEEQKKRWLPGIADGTLTMAFAITEPDAGSNSHKITT
MAB_2065c|M.abscessus_ATCC_199 PTICG-TIIQAFGSEELKQQWLPGIASGETIMAFAITEPDAGSNSHNIST
MSMEG_4836|M.smegmatis_MC2_155 MSIFGMQPVVVFGSDEMKAETLPRIVDGDLHVCFGVTEPGAGLDTSRITT
TH_4562|M.thermoresistible__bu LSIFGMQPVVLFGSDEMKAATLPRIVDGSLHVCFGVTEPGAGLDTTRITT
Mflv_2442|M.gilvum_PYR-GCK LSIFGMQPVVKHGSDELKAATLPRIVDGDLHVCFGVTEPGAGLDTSRITT
Mvan_4210|M.vanbaalenii_PYR-1 LSIFGMQPVVKHGSDELKAATLPRIVNGDLHVCFGVTEPGAGLDTSRITT
MMAR_4741|M.marinum_M LSIFGMQPVVVHGSDELKARTLPSVATGETHVCFGVTEPGAGLDTSRITT
MLBr_00737|M.leprae_Br4923 --KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRT
* : *::: * ** :. .:.::* ** :: : *
Mb0997c|M.bovis_AF2122/97 TARRDGSD----WIIKGQKVFISGIDQAQAVLVVGRSEEAKTGKLR----
Rv0972c|M.tuberculosis_H37Rv TARRDGSD----WIIKGQKVFISGIDQAQAVLVVGRSEEAKTGKLR----
MUL_4707|M.ulcerans_Agy99 TARRDGSD----WIIKGQKVYISGIDQAQAVLVVGRTEEAKTGKLR----
MAV_1088|M.avium_104 TARRDGSD----WILKGQKVFISGIDQAQAVLVVARTEEAKTGKLR----
MAB_2065c|M.abscessus_ATCC_199 TAKRDGDD----WVINGTKYYISGVDEAEAILVVTRTSTNDSGRGL----
MSMEG_4836|M.smegmatis_MC2_155 FARRDGDHPDADYVVNGRKVWISKALESEKILLLTRTESREDVEKRGGKP
TH_4562|M.thermoresistible__bu FARREGDH----YVVNGRKVWISKAMESEKILLLARTEPLDAAERPGTKK
Mflv_2442|M.gilvum_PYR-GCK FAKREGDH----YRVNGRKVWISKALESEKILLLTRTETPEDLKQRGAKK
Mvan_4210|M.vanbaalenii_PYR-1 FAKREGDK----YRVNGRKVWISKALESEKILLLTRTESPDDLEKRGAKK
MMAR_4741|M.marinum_M FAKRDGDR----YVVNGRKVWISKAVESDKILLLTRTQSYDEVK----KK
MLBr_00737|M.leprae_Br4923 RAKADGDD----WILNGFKCWITNGGKSTWYTVMAVTDPDKGANG-----
*: :*. : ::* * :*: :: :: :. . .
Mb0997c|M.bovis_AF2122/97 ---PALFVVPTDAPGFSYTP-IEMELVSPERQFQVFLDDVRLPADALVGA
Rv0972c|M.tuberculosis_H37Rv ---PALFVVPTDAPGFSYTP-IEMELVSPERQFQVFLDDVRLPADALVGA
MUL_4707|M.ulcerans_Agy99 ---PALFVVPTDTPGFTYTA-IDMELVSPERQFQVFLDDVRLPADALVGA
MAV_1088|M.avium_104 ---PALFVVPTDAPGFSATP-IEMELISPERQFQVFLDDVRLPSDALVGA
MAB_2065c|M.abscessus_ATCC_199 ---LSLLVVPTDVPGLTKSL-IPVQAVTPEKQFTLFFDDVRVPAANLIGT
MSMEG_4836|M.smegmatis_MC2_155 TEGLSLFLTDLDRDHVDIRP-INKMGRNAVSSNELFIDDLRVPVEHRIGE
TH_4562|M.thermoresistible__bu TDGLSLFLTDLDRDHVDIRP-IKKMGRNAVGSNELFIDDLRVPVEDRIGE
Mflv_2442|M.gilvum_PYR-GCK TDGMTLFLTDLDRDHVDIRP-IRKMGRNAVSSNELFIDDLMVPVEHRVGE
Mvan_4210|M.vanbaalenii_PYR-1 TDGMTLFLTDLDRNHVDIRP-IRKMGRNAVSSNEVFIDDLMVPVEHRVGE
MMAR_4741|M.marinum_M TDGMTLFITDLDRSRVEVRP-IRKMGRNAVSSNELFIDNLEIPVEDRIGE
MLBr_00737|M.leprae_Br4923 ---ISAFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGE
: ::. * . . :::*. :* :*
Mb0997c|M.bovis_AF2122/97 EDAAIAHLFAGLNPERIMGAASAVGMGRFALGRAVDYVKTRKVWSTPIGA
Rv0972c|M.tuberculosis_H37Rv EDAAIAQLFAGLNPERIMGAASAVGMGRFALGRAVDYVKTRKVWSTPIGA
MUL_4707|M.ulcerans_Agy99 QDAAIAQLFAGLNPERIMGAASAVGMGRFALNRAVDYVKTRQVWSTPIGA
MAV_1088|M.avium_104 EDAAIAQLFAGLNPERIMGAASSVGMGRFALARAAEYVKTRQVWGTPVGA
MAB_2065c|M.abscessus_ATCC_199 ENEGLRQVFMGLNPERIMGAALGNGIGRYALDKAADYARNRHVWGAPIGS
MSMEG_4836|M.smegmatis_MC2_155 EGKGFGYILHGLNPERMLIAAEALGIGRVALDRAVKYANDRVVFDRPIGM
TH_4562|M.thermoresistible__bu EGKGFSYILHGLNPERMLIAAEALGIGRVALDRAVKYANERHVFDRPIGM
Mflv_2442|M.gilvum_PYR-GCK EGQGFKYILDGLNPERMLIAAEALGIGRVALEKAVKYGNERVVFNRPIGM
Mvan_4210|M.vanbaalenii_PYR-1 EGKGFKYILDGLNPERMLIAAEALGIGRVALEKAVKYGNERHVFNRPIGM
MMAR_4741|M.marinum_M EGNGFQYILDGLNPERMLIAAEALGIGRVALDKAVKYGNEREVFGRPIGM
MLBr_00737|M.leprae_Br4923 PGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESIST
. .: : *: * .* . *:.: ** * * . * :. .:.
Mb0997c|M.bovis_AF2122/97 HQGLAHPLAQCHIEVELAKLMTQKAATLYDHGDDFGAAEAANMAKYAAAE
Rv0972c|M.tuberculosis_H37Rv HQGLAHPLAQCHIEVELAKLMTQKAATLYDHGDDFGAAEAANMAKYAAAE
MUL_4707|M.ulcerans_Agy99 HQGLAHPLAQCHIEVELAKLMMQKAATLYDHGDDGGAAEAANMAKYAAAE
MAV_1088|M.avium_104 HQGLSHPLAQCHIEVELAKLMMQKAATLYDAGNDMGAAEAANMAKYAAAE
MAB_2065c|M.abscessus_ATCC_199 HQGVSHPLAHAKIQVELARLMTQRAAALHDAGDP-GSGEASNMAKYAAAE
MSMEG_4836|M.smegmatis_MC2_155 NQGIQFPLADSLARLDAAELVLRKATWLYDNGKP--CGREANMAKYLCAD
TH_4562|M.thermoresistible__bu NQGIQFPLADSLARLDAAELVLRKATWLYDNGKP--CGREANMAKYLCAD
Mflv_2442|M.gilvum_PYR-GCK NQGLQFPLADSLARLDAAELVLRKATWLYDNGKP--CGREANTAKYLCAD
Mvan_4210|M.vanbaalenii_PYR-1 NQGLQFPLADSLARLDAAELVLRKATWLYDNGKP--CGREANTAKYLCAD
MMAR_4741|M.marinum_M NQGLQFPLADSLARLDAAELMLRKATWLYDNGKP--CGREANTAKYLCAD
MLBr_00737|M.leprae_Br4923 FQSIQFMLADMAMKVEAARLIVYAAAARAERGEP-DLGFISAASKCFASD
*.: . **. .:: *.*: *: : *. . : :* .::
Mb0997c|M.bovis_AF2122/97 ASSRAVDQAVQSMGGNGLTKEYGVAAMMTSARLARIAPISREMVLNFVAQ
Rv0972c|M.tuberculosis_H37Rv ASSRAVDQAVQSMGGNGLTKEYGVAAMMTSARLARIAPISREMVLNFVAQ
MUL_4707|M.ulcerans_Agy99 ASARSVDQAVQSMGGNGLTQEYGVAAMLTSSRLARIAPISREMVLNFVAQ
MAV_1088|M.avium_104 AATRSVDQAVQSMGGNGLTKEYGVAAMLASARLGRIAPVSREMVLNFVAQ
MAB_2065c|M.abscessus_ATCC_199 AGILALDQAIQTHGGNGMATEYGLATLWGPARLMRTAPISREMILNFVAQ
MSMEG_4836|M.smegmatis_MC2_155 AGFAAADRALQTHGGMGYSEEYNISRFFRESRLMKIAPVSQEMILNFLGA
TH_4562|M.thermoresistible__bu AGFAAADRALQTHGGMGYSEEYSISRFFRESRLMKIAPVSQEMILNYLGS
Mflv_2442|M.gilvum_PYR-GCK AGFGAADRALQLHGGMGYSEEYHVSRYFRESRLMKIAPVSQEMILNFLGE
Mvan_4210|M.vanbaalenii_PYR-1 AGFGAADRALQLHGGMGYSEEYHVSRYFRESRLMKIAPVSQEMILNFLGE
MMAR_4741|M.marinum_M AGFAAADRALQTHGGMGYAEEYHVARYFREARLMKIAPVSQEMILNFLGS
MLBr_00737|M.leprae_Br4923 IAMEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSR
. *:* ** * : :: : ::: : :.:: :.
Mb0997c|M.bovis_AF2122/97 TSLGLPRSY
Rv0972c|M.tuberculosis_H37Rv TSLGLPRSY
MUL_4707|M.ulcerans_Agy99 TSLGLPRSY
MAV_1088|M.avium_104 TSLGLPRSY
MAB_2065c|M.abscessus_ATCC_199 HSLKLPASY
MSMEG_4836|M.smegmatis_MC2_155 NVLGLPKSY
TH_4562|M.thermoresistible__bu HVLGLPRSY
Mflv_2442|M.gilvum_PYR-GCK HVLGLPRSY
Mvan_4210|M.vanbaalenii_PYR-1 HVLGLPRSY
MMAR_4741|M.marinum_M HVLKLPRSY
MLBr_00737|M.leprae_Br4923 ALLR-----
*