For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDTVTVVRAGPPETDDTTTPDSPTVVERDFRPSASESRRGRTRRLSVGIRFVVPVLLLVAWQVAAAWELV SPSVLPGPLTILQSYRELWQIGDLQAALPISLQRAGLGLLIGGITGLVLGVAAGLWAVAERIYDAPLQML RTIPFIAMVPLFVVWFGIGEESKVALIVGASIFPVYLNTYHGIRAIDRKLLEVGQTFALSRRETIRLVVV PLAMPSILVGWRYAAGTALLALVAAEQINTTSGIGYILNTANQFQRTDIIIAGILVYAALGILVDVIMRL IERAALPWRATHDGH*
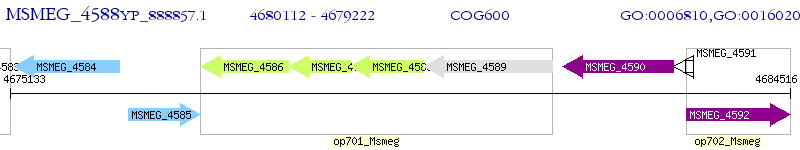
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4588 | - | - | 100% (296) | ABC nitrate/sulfonate/bicarbonate family protein transporter, inner membrane subunit |
| M. smegmatis MC2 155 | MSMEG_3854 | - | 3e-58 | 45.35% (258) | putative aliphatic sulfonates transport permease protein SsuC |
| M. smegmatis MC2 155 | MSMEG_5530 | - | 2e-46 | 37.96% (245) | putative aliphatic sulfonates transport permease protein SsuC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3783c | proW | 7e-12 | 25.00% (200) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3159 | - | 2e-42 | 36.67% (240) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3757c | proW | 7e-12 | 25.00% (200) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2216 | - | 2e-60 | 43.68% (261) | sulfonate ABC transporter, permease protein SsuC |
| M. marinum M | MMAR_0033 | - | 1e-16 | 32.43% (148) | hypothetical protein MMAR_0033 |
| M. avium 104 | MAV_2433 | - | 2e-56 | 45.23% (241) | putative aliphatic sulfonates transport permease protein SsuC |
| M. thermoresistible (build 8) | TH_4654 | - | 2e-48 | 37.04% (243) | PUTATIVE putative binding-protein-dependent transport systems |
| M. ulcerans Agy99 | MUL_0032 | - | 6e-13 | 33.90% (118) | hypothetical protein MUL_0032 |
| M. vanbaalenii PYR-1 | Mvan_3413 | - | 1e-53 | 42.18% (275) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3783c|M.bovis_AF2122/97 --------------------------------------------------
Rv3757c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2433|M.avium_104 MSIATALDAAPKRLGGAASG-------------APGG--LWARRKWEV-V
Mvan_3413|M.vanbaalenii_PYR-1 MTRPADPTAP---LAQANSG-------------RVG---RRADRRWAL-I
MAB_2216|M.abscessus_ATCC_1997 MTTRNTLVAAAATDSDAPES-------------GPSRREWVLERRWEI-V
MSMEG_4588|M.smegmatis_MC2_155 MTDTVTVVRAGPPETDDTTTPDSPTVVERDFRPSASESRRGRTRRLSVGI
Mflv_3159|M.gilvum_PYR-GCK MTAVAAPLTTP--RGPAAPAPELVDVIAG----HRGR-RSIWG-RIPRPV
TH_4654|M.thermoresistible__bu VTSLQTARPAPRLRSPAQRRADVTDAQLL----HRGEHRTRLGPGRAVPA
MMAR_0033|M.marinum_M MSVAGNDINVEGAVKGGAEG------------------------PPTAPT
MUL_0032|M.ulcerans_Agy99 MGAAATHV----CLRGGR----------------------------SAGL
Mb3783c|M.bovis_AF2122/97 -------MHYLMTHPGAAWALT------------------------VVHL
Rv3757c|M.tuberculosis_H37Rv -------MHYLMTHPGAAWALT------------------------VVHL
MAV_2433|M.avium_104 RLVS-PLALLALWQLGSAIGVIAQDVLPAPSLILQAGVELTRNGQLADAL
Mvan_3413|M.vanbaalenii_PYR-1 RLAS-PFILLAVWQLGSAVGLIPQDVLPAPSLIAEAGIELIGNGQLVDAL
MAB_2216|M.abscessus_ATCC_1997 RFLS-PLAVLLIWQAGSAWGLIDPDILPAPSTIARAGFELIANGQLAEAL
MSMEG_4588|M.smegmatis_MC2_155 RFVV-PVLLLVAWQVAAAWELVSPSVLPGPLTILQSYRELWQIGDLQAAL
Mflv_3159|M.gilvum_PYR-GCK RRMAGPVAILAVWTIGSTSGLLNTDLFPPPSDVAVTAWHLLTNGGLALHV
TH_4654|M.thermoresistible__bu ALAVGPLLLVALWLITSQLGILDPRTLPHPFDIARTAAELWSDGRLPANV
MMAR_0033|M.marinum_M TPGRPSVGRWALQPLTCAFAVAGALAYVTVADVSESERRSLSVSHLLQLL
MUL_0032|M.ulcerans_Agy99 CDGGRCVR--------VGAPLAERVASVAAAALTPGDQ---PVGHGAHVC
:
Mb3783c|M.bovis_AF2122/97 RLSLLPVLIGLMSAVPLGLLVQRAPLLRRLT--TATASVIFTIP-SLALF
Rv3757c|M.tuberculosis_H37Rv RLSLLPVLIGLMSAVPLGLLVQRAPLLRRLT--TATASVIFTIP-SLALF
MAV_2433|M.avium_104 HISTVRVIEGLALGGVIGIVAGAAVGLSRWV--EATVDPPLQMIRALPHL
Mvan_3413|M.vanbaalenii_PYR-1 TVSGLRVVEGLILGGVIGIALGTAVGLSKWF--EATVDPPMQMVRALPHL
MAB_2216|M.abscessus_ATCC_1997 RVSGVRAAEGLLLGGVIGVVLGAAVGLSRLA--DAVVDPTMQMIRALPHL
MSMEG_4588|M.smegmatis_MC2_155 PISLQRAGLGLLIGGITGLVLGVAAGLWAVA--ERIYDAPLQMLRTIPFI
Mflv_3159|M.gilvum_PYR-GCK GTSATRVLIGTALGITVGVVLAVLAGLTRTG--EDLLDWSMQILKAVPNF
TH_4654|M.thermoresistible__bu AASLKLSLISLAIGVAAGLALALLAGLSRIG--EALVDGPIQIKRAVPTL
MMAR_0033|M.marinum_M REHLVISLSATVLTCVVAIPVGIALTRGSMRRYSKPIITVAGFGQAAPAI
MUL_0032|M.ulcerans_Agy99 GGD---SGGNRIDARTEAALFEADHHRRQFR------SGRAGYRPDR---
.
Mb3783c|M.bovis_AF2122/97 VVLPLIIGTRILDEANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATA
Rv3757c|M.tuberculosis_H37Rv VVLPLIIGTRILDEANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATA
MAV_2433|M.avium_104 GLIPLFILWFGIGELPKVLLVALGVVFPLYLNTFSAIRQVDPKMLETAQV
Mvan_3413|M.vanbaalenii_PYR-1 GLIPLFIIWFGIGELPKVLLVALGVSFPLYLNTFSAIRQVDPKLFETAQV
MAB_2216|M.abscessus_ATCC_1997 GLIPLFIVWFGIGELPKVLMVALGAAFPLYLNTTSAIRQVDPKLFETAQV
MSMEG_4588|M.smegmatis_MC2_155 AMVPLFVVWFGIGEESKVALIVGASIFPVYLNTYHGIRAIDRKLLEVGQT
Mflv_3159|M.gilvum_PYR-GCK ALTPLLIIWMGIGEGPKIVLITMGVAIAVYINTYSGIRGVDQQLVEMAQT
TH_4654|M.thermoresistible__bu AIIPLAIIWFGIGDTMKIVIIATSVLIPVYINTTAQLKGVDLRHVELAQT
MMAR_0033|M.marinum_M GLIALGAVVFGIGQLGAIVALTVYGAMPIIANTVTGLDGVDRRIVEAARG
MUL_0032|M.ulcerans_Agy99 ---AGGGRVRDRSARRDRCPHRLR-RDPIIANTVTGLDGVDRRIVEAARG
. . : . : : : : .
Mb3783c|M.bovis_AF2122/97 IGYSRIAQMLKVELPLSIPVLVAGLR-VVAVTNIAMVSVGSVIGIGGLGT
Rv3757c|M.tuberculosis_H37Rv IGYSRIAQMLKVELPLSIPVLVAGLR-VVAVTNIAMVSVGSVIGIGGLGT
MAV_2433|M.avium_104 LGFSFFQRFRRILVPGTAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGF
Mvan_3413|M.vanbaalenii_PYR-1 LGFSFWQRFRSIIVPSAAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGF
MAB_2216|M.abscessus_ATCC_1997 LGFSFWQRLRVIVIPGATPQVLVGLRQSLALSWLTLIVAEQINADAGVGF
MSMEG_4588|M.smegmatis_MC2_155 FALSRRETIRLVVVPLAMPSILVGWRYAAGTALLALVAAEQINTTSGIGY
Mflv_3159|M.gilvum_PYR-GCK LEARRGTLITQVILPGAMPNFLVGLRLGLSSAWLSLIFAEMINTTEGIGF
TH_4654|M.thermoresistible__bu VGLSRVQFIRKVALPGALPGFFTGLRLAVTISWTALVVVEQVNATSGVGF
MMAR_0033|M.marinum_M MGLSAFSTLRRVELPLSLPVIVAGVR-TALVLIVGTAALASFTGGGGLGQ
MUL_0032|M.ulcerans_Agy99 MGLSAFSTLRRVELPLSLPVIVAGVR-TALVLIVGTAALASFTGGRGLGQ
. : : :* : * ...* * . *:*
Mb3783c|M.bovis_AF2122/97 WFTAGYQTNKSDQIVAGVVAMFLLAIVVDVVINLAGRLATPWERAPRAAR
Rv3757c|M.tuberculosis_H37Rv WFTAGYQTNKSDQIVAGVVAMFLLAIVVDVVINLAGRLATPWERAPRAAR
MAV_2433|M.avium_104 LINNARDFLRIDIIIFGLTIYALLGIITDAVVRLIERRAVRYRH------
Mvan_3413|M.vanbaalenii_PYR-1 LIMNARDFLRIDIIIFGLIVYALLGIGTDAVVRALERRALRYRS------
MAB_2216|M.abscessus_ATCC_1997 LINNARDFLRIDVIIFGLVVYALLGIVTDAIVRVLERRALRYRS------
MSMEG_4588|M.smegmatis_MC2_155 ILNTANQFQRTDIIIAGILVYAALGILVDVIMRLIERAALPWRATHDGH-
Mflv_3159|M.gilvum_PYR-GCK LMSRAQTNLQFDVSLLVIVIYAVLGLASYALVRLLERLLLSWRNGFAGFE
TH_4654|M.thermoresistible__bu LMTQARLYGQLEVVVVGLVVYAVFGLAGDHLVRTIERRALSWRRALGG--
MMAR_0033|M.marinum_M LITTGIKLQQPVTLIVGAILVAALALFIDWLARLIEMVAAPRGL------
MUL_0032|M.ulcerans_Agy99 LITTGIKLQQPVTLIVGAILVAALALFIDWLARLIEMVAAPRGL------
: . : : :.: : .
Mb3783c|M.bovis_AF2122/97 RRRQVAAPITGGAR
Rv3757c|M.tuberculosis_H37Rv RRRQVAAPITGGAR
MAV_2433|M.avium_104 --------------
Mvan_3413|M.vanbaalenii_PYR-1 --------------
MAB_2216|M.abscessus_ATCC_1997 --------------
MSMEG_4588|M.smegmatis_MC2_155 --------------
Mflv_3159|M.gilvum_PYR-GCK ATA-----------
TH_4654|M.thermoresistible__bu --------------
MMAR_0033|M.marinum_M --------------
MUL_0032|M.ulcerans_Agy99 --------------