For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTIAGTRNGPTTAGEVSWQRRCAPPITAILIAIALWWSATSVLSGPESVLRQTAPQHVVPALSGLLERQV LLGDLGVSLWRLSIGLAVATVIGMPLGLLIGSSRIAERAGGPIIAFLRMISPLSWTPIALAVFGIGNQPV VFLVAVAAVWPVLLNTAAGVRAVDPGLLNVARSFHATRWEMMLAVILPAIRTHVQTGIRLALGVAWIVLV PAEMLGVRSGLGYQILNARDQLAYDQVVAVIVVIGILGFLLDTTARWLLNPARRA
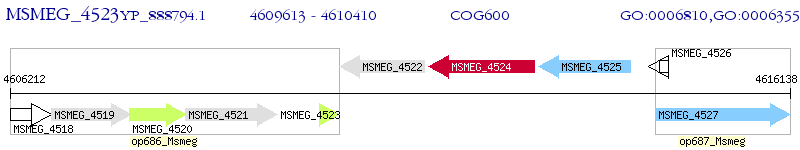
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4523 | - | - | 100% (265) | binding-protein-dependent transport systems inner membrane component |
| M. smegmatis MC2 155 | MSMEG_3854 | - | 2e-31 | 32.55% (255) | putative aliphatic sulfonates transport permease protein SsuC |
| M. smegmatis MC2 155 | MSMEG_1161 | - | 7e-28 | 32.62% (233) | taurine transport system permease protein TauC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3782c | proZ | 8e-09 | 24.53% (212) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3159 | - | 5e-24 | 29.55% (247) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3756c | proZ | 8e-09 | 24.53% (212) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2463 | - | 7e-94 | 68.92% (251) | putative sulfonate ABC transporter, permease |
| M. marinum M | MMAR_0035 | - | 4e-13 | 30.68% (176) | ABC transporter permease |
| M. avium 104 | MAV_3399 | - | 7e-31 | 32.81% (256) | ABC transporter permease protein |
| M. thermoresistible (build 8) | TH_4654 | - | 3e-20 | 28.45% (239) | PUTATIVE putative binding-protein-dependent transport systems |
| M. ulcerans Agy99 | MUL_0034 | - | 7e-13 | 30.68% (176) | ABC transporter permease |
| M. vanbaalenii PYR-1 | Mvan_3413 | - | 2e-30 | 33.33% (234) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4523|M.smegmatis_MC2_155 -------------MTIAGTRNGPTTAGEVSWQR-----RCAPPITAILIA
MAB_2463|M.abscessus_ATCC_1997 -------------MSIIERAPRAVPVGTVAAQKALLLSRIWPPALAIAAT
Mflv_3159|M.gilvum_PYR-GCK MTAVAAPLTTP--RGPAAPAPELVDVIAGHRGR-RSIWG-RIPRPVRRMA
TH_4654|M.thermoresistible__bu VTSLQTARPAPRLRSPAQRRADVTDAQLLHRGEHRTRLGPGRAVPAALAV
Mvan_3413|M.vanbaalenii_PYR-1 MT------------RPADPTAPLAQANSGRVGR-----RADRRWALIRLA
Mb3782c|M.bovis_AF2122/97 --------------------------------------------MNFLQQ
Rv3756c|M.tuberculosis_H37Rv --------------------------------------------MNFLQQ
MMAR_0035|M.marinum_M -------------------------------------------MEAATEF
MUL_0034|M.ulcerans_Agy99 --------------------------------------------------
MAV_3399|M.avium_104 ---------MTAQVVADQVVGGVISAPPLARPRRGTLRWQSRVLRVVSVA
MSMEG_4523|M.smegmatis_MC2_155 IALWWSATSVLSGPESVLR---QTAPQHVVPALSGLLERQVLLGDLGVSL
MAB_2463|M.abscessus_ATCC_1997 VAVWWLASSVLSQPYSLLR---QTAPDKAFRAAMDLLNRGVLLQDAGISL
Mflv_3159|M.gilvum_PYR-GCK GPVAILAVWTIGSTSGLLNTDLFPPPSDVAVTAWHLLTNGGLALHVGTSA
TH_4654|M.thermoresistible__bu GPLLLVALWLITSQLGILDPRTLPHPFDIARTAAELWSDGRLPANVAASL
Mvan_3413|M.vanbaalenii_PYR-1 SPFILLAVWQLGSAVGLIPQDVLPAPSLIAEAGIELIGNGQLVDALTVSG
Mb3782c|M.bovis_AF2122/97 ALSYLLTASNWTGPV-------------------------GLAVRTCEHL
Rv3756c|M.tuberculosis_H37Rv ALSYLLTASNWTGPV-------------------------GLAVRTCEHL
MMAR_0035|M.marinum_M MGLYDFVVDRWS----------------------------VLWFLAYQHI
MUL_0034|M.ulcerans_Agy99 MGLYDFVVDRWS----------------------------VLWFLAYQHI
MAV_3399|M.avium_104 AAIGLWQLLTADKVRLLLRFDTLPTVTEIVGALHRRLAAGEYWLDLAQSL
MSMEG_4523|M.smegmatis_MC2_155 WRLSIGLAVATVIGMPLGLLIGSSRIAERAGGPIIAFLRMISPLSWTPIA
MAB_2463|M.abscessus_ATCC_1997 WRLLIGLCLAAVIGVPAGLLLGVSTTAERAARPVIQFLRMISPLSWTPIA
Mflv_3159|M.gilvum_PYR-GCK TRVLIGTALGITVGVVLAVLAGLTRTGEDLLDWSMQILKAVPNFALTPLL
TH_4654|M.thermoresistible__bu KLSLISLAIGVAAGLALALLAGLSRIGEALVDGPIQIKRAVPTLAIIPLA
Mvan_3413|M.vanbaalenii_PYR-1 LRVVEGLILGGVIGIALGTAVGLSKWFEATVDPPMQMVRALPHLGLIPLF
Mb3782c|M.bovis_AF2122/97 EYTAVAVAASALIAVPVGLLIGHTGRGTLLVVGAVNGLRALPTLGVLLLG
Rv3756c|M.tuberculosis_H37Rv EYTAVAVAASALIAVPVGLLIGHTGRGTLLVVGAVNGLRALPTLGVLLLG
MMAR_0035|M.marinum_M SLVGQTLILATAIALILGVLIYNSPLGVAAGNAVTAVGLTIPSYALLGVL
MUL_0034|M.ulcerans_Agy99 SLVGQALILATAIALILGVLIYNSPLRVAAGNAVTAVGLTIPSYALLGVL
MAV_3399|M.avium_104 LRILTGFGLAAVIGVATGVLLGRSRLFADVFGPLAELARPIPAIAMVPVA
. .: . : :. . :
MSMEG_4523|M.smegmatis_MC2_155 LAVFGIGNQPVVFLVAVAAVWPVLLNTAAGVRAVDPGLLNVARSFHATRW
MAB_2463|M.abscessus_ATCC_1997 VAVFGIGNQPVVFLIATAAVWPILLNTTAGVHSIEPGYLHVARSFHATRS
Mflv_3159|M.gilvum_PYR-GCK IIWMGIGEGPKIVLITMGVAIAVYINTYSGIRGVDQQLVEMAQTLEARRG
TH_4654|M.thermoresistible__bu IIWFGIGDTMKIVIIATSVLIPVYINTTAQLKGVDLRHVELAQTVGLSRV
Mvan_3413|M.vanbaalenii_PYR-1 IIWFGIGELPKVLLVALGVSFPLYLNTFSAIRQVDPKLFETAQVLGFSFW
Mb3782c|M.bovis_AF2122/97 VLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMTES
Rv3756c|M.tuberculosis_H37Rv VLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMTES
MMAR_0035|M.marinum_M VGIVGLGVLPSVIMLTFFGVFPILRNVVVGLTGVDKGLVESARGMGMSRL
MUL_0034|M.ulcerans_Agy99 VGIVGLGVLPSVIMPTFFGVFPILRNVVVGLTGVDKGLVESARGMGMSRL
MAV_3399|M.avium_104 ILLFPTDEAGIVFITFLAAYFPIMVSTRHAVRALPTLWEDSVRTLGGGRW
: . . :. .: .. : : . .: .
MSMEG_4523|M.smegmatis_MC2_155 EMMLAVILPAIRTHVQTGIRLALGVAWIVLVPAEMLGVRSGLGYQILNAR
MAB_2463|M.abscessus_ATCC_1997 EVLTAVILPAIRGHIQTGIRVALGVAWVVLVPAEMLGVRSGLGYQVLNAR
Mflv_3159|M.gilvum_PYR-GCK TLITQVILPGAMPNFLVGLRLGLSSAWLSLIFAEMINTTEGIGFLMSRAQ
TH_4654|M.thermoresistible__bu QFIRKVALPGALPGFFTGLRLAVTISWTALVVVEQVNATSGVGFLMTQAR
Mvan_3413|M.vanbaalenii_PYR-1 QRFRSIIVPSAAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGFLIMNAR
Mb3782c|M.bovis_AF2122/97 QVLLRVEVPNALPLMLGGLRSATLQVVATATVAAYASLGGLGGYLIDGIK
Rv3756c|M.tuberculosis_H37Rv QVLLRVEVPNALPLMLGGLRSATLQVVATATVAAYASLGGLGGYLIDGIK
MMAR_0035|M.marinum_M TTLVRLELPLAWPVIMTGVRVSAQMLMGIAAIVAFALGPGLGGYIFSGIS
MUL_0034|M.ulcerans_Agy99 TTLVRLELPLAWPVIMTGVRVSAQMLMGIAAIVAFALGPGLGGYIFSGIS
MAV_3399|M.avium_104 QVLTQVVLPGILPGVFGGLSVGMGVAWICVISAEMISGRLGVGYRTWQDY
: : :* . *: . . *:
MSMEG_4523|M.smegmatis_MC2_155 D-------QLAYDQVVAVIVVIGILGFLLDTTAR---WLLNPARRA----
MAB_2463|M.abscessus_ATCC_1997 D-------QLAYDQVVAVILVIGILGYALDMAAK---RLLASRFRVPSAG
Mflv_3159|M.gilvum_PYR-GCK T-------NLQFDVSLLVIVIYAVLGLASYALVR---LLERLLLSWRNGF
TH_4654|M.thermoresistible__bu L-------YGQLEVVVVGLVVYAVFGLAGDHLVR---TIERRALSWRRAL
Mvan_3413|M.vanbaalenii_PYR-1 D-------FLRIDIIIFGLIVYALLGIGTDAVVR---ALERRALRYRS--
Mb3782c|M.bovis_AF2122/97 ERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKLAAVVDKPA
Rv3756c|M.tuberculosis_H37Rv ERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKLAAVVDKPA
MMAR_0035|M.marinum_M R------MGGANATNSIVAATIGILILAVILDT-----VLNLIARLTTPR
MUL_0034|M.ulcerans_Agy99 R------MGGANATNSIVAATIGILILAVILDT-----VLNVIARLTTPR
MAV_3399|M.avium_104 T-------VLAYPQVFVGIITIGVLGFATSAAVELVGRRVTRWLPRAQDG
.:: .
MSMEG_4523|M.smegmatis_MC2_155 --------
MAB_2463|M.abscessus_ATCC_1997 --------
Mflv_3159|M.gilvum_PYR-GCK AGFEATA-
TH_4654|M.thermoresistible__bu GG------
Mvan_3413|M.vanbaalenii_PYR-1 --------
Mb3782c|M.bovis_AF2122/97 AGGGHALR
Rv3756c|M.tuberculosis_H37Rv AGGGHALR
MMAR_0035|M.marinum_M GIRA----
MUL_0034|M.ulcerans_Agy99 EIRA----
MAV_3399|M.avium_104 AR------