For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVIPPAEPQAGRGRRFRALTWRLVRPVVVIALFLGVWETAPRIGMVDNVFLPPLSHVVQAWFTLLGNGQL WEHVSASLSRALAGFALAILVSIPLGVAIGWYRPVADFLNPILELFRNTAALALLPVFVLILGIGETSKV ALVVYASAFPILLNTISGVRTVDPLLIKSARSLGLSPVRLFQKVVLPAAVPTIFTGLRMAAASSILVLIA AEMVGAKAGLGYLITASQLNFQIPNMYAGIVTIALVGVVFNSVLVAIEGRLSGWRNTN
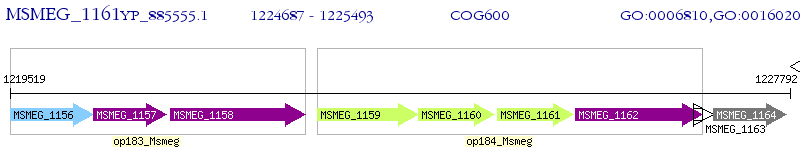
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1161 | - | - | 100% (268) | taurine transport system permease protein TauC |
| M. smegmatis MC2 155 | MSMEG_3854 | - | 5e-39 | 32.94% (252) | putative aliphatic sulfonates transport permease protein SsuC |
| M. smegmatis MC2 155 | MSMEG_5530 | - | 7e-36 | 33.73% (255) | putative aliphatic sulfonates transport permease protein SsuC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3782c | proZ | 1e-11 | 27.47% (182) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3159 | - | 2e-35 | 34.50% (258) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3756c | proZ | 1e-11 | 27.47% (182) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2216 | - | 6e-41 | 35.63% (261) | sulfonate ABC transporter, permease protein SsuC |
| M. marinum M | MMAR_0033 | - | 2e-15 | 27.75% (191) | hypothetical protein MMAR_0033 |
| M. avium 104 | MAV_2433 | - | 1e-38 | 35.77% (246) | putative aliphatic sulfonates transport permease protein SsuC |
| M. thermoresistible (build 8) | TH_4654 | - | 8e-31 | 29.75% (242) | PUTATIVE putative binding-protein-dependent transport systems |
| M. ulcerans Agy99 | MUL_4374 | proZ | 1e-13 | 31.02% (187) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_3413 | - | 3e-39 | 34.21% (266) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3159|M.gilvum_PYR-GCK MTAVAAPLTTP--RGPAAPAPELVDVIAGHRGR-RSIWG-RIPRPVRRMA
TH_4654|M.thermoresistible__bu VTSLQTARPAPRLRSPAQRRADVTDAQLLHRGEHRTRLGPGRAVPAALAV
MAV_2433|M.avium_104 MSIATALDAAPKRLGGAA-----------SGAPGG--LWARRKWEVVRLV
Mvan_3413|M.vanbaalenii_PYR-1 MTRPADPTAP---LAQAN-----------SGRVG---RRADRRWALIRLA
MAB_2216|M.abscessus_ATCC_1997 MTTRNTLVAAAATDSDAP-----------ESGPSRREWVLERRWEIVRFL
MSMEG_1161|M.smegmatis_MC2_155 ---MVIPPAEPQAGRGRR-------------------FRALTWRLVRPVV
Mb3782c|M.bovis_AF2122/97 --------------------------------------------------
Rv3756c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4374|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0033|M.marinum_M MSVAGNDINVEGAVKGGAE--------------------GPPTAPTTPGR
Mflv_3159|M.gilvum_PYR-GCK GPVAILAVWTIGSTSGLLNTDLFPPPSDVAVTAWHLLTNGGLALHVGTSA
TH_4654|M.thermoresistible__bu GPLLLVALWLITSQLGILDPRTLPHPFDIARTAAELWSDGRLPANVAASL
MAV_2433|M.avium_104 SPLALLALWQLGSAIGVIAQDVLPAPSLILQAGVELTRNGQLADALHIST
Mvan_3413|M.vanbaalenii_PYR-1 SPFILLAVWQLGSAVGLIPQDVLPAPSLIAEAGIELIGNGQLVDALTVSG
MAB_2216|M.abscessus_ATCC_1997 SPLAVLLIWQAGSAWGLIDPDILPAPSTIARAGFELIANGQLAEALRVSG
MSMEG_1161|M.smegmatis_MC2_155 VIALFLGVWETAPRIGMVDNVFLPPLSHVVQAWFTLLGNGQLWEHVSASL
Mb3782c|M.bovis_AF2122/97 -------------------MNFLQQALSYLLTASNWTGPVGLAVRTCEHL
Rv3756c|M.tuberculosis_H37Rv -------------------MNFLQQALSYLLTASNWTGPVGLAVRTCEHL
MUL_4374|M.ulcerans_Agy99 -------------------MNFLQQALSYLSTAANWTGSAGLAHRTAEHL
MMAR_0033|M.marinum_M PSVGRWALQPLTCAFAVAGALAYVTVADVSESERRSLSVSHLLQLLREHL
: *
Mflv_3159|M.gilvum_PYR-GCK TRVLIGTALGITVGVVLAVLAG--LTRTGEDLLDWSMQILKAVPNFALTP
TH_4654|M.thermoresistible__bu KLSLISLAIGVAAGLALALLAG--LSRIGEALVDGPIQIKRAVPTLAIIP
MAV_2433|M.avium_104 VRVIEGLALGGVIGIVAGAAVG--LSRWVEATVDPPLQMIRALPHLGLIP
Mvan_3413|M.vanbaalenii_PYR-1 LRVVEGLILGGVIGIALGTAVG--LSKWFEATVDPPMQMVRALPHLGLIP
MAB_2216|M.abscessus_ATCC_1997 VRAAEGLLLGGVIGVVLGAAVG--LSRLADAVVDPTMQMIRALPHLGLIP
MSMEG_1161|M.smegmatis_MC2_155 SRALAGFALAILVSIPLGVAIG--WYRPVADFLNPILELFRNTAALALLP
Mb3782c|M.bovis_AF2122/97 EYTAVAVAASALIAVPVGLLIG--HTGRGTLLVVGAVNGLRALPTLGVLL
Rv3756c|M.tuberculosis_H37Rv EYTAVAVAASALIAVPVGLLIG--HTGRGTLLVVGAVNGLRALPTLGVLL
MUL_4374|M.ulcerans_Agy99 EYTALAATASALVAIPIGLIIG--HTGRGQLLVVSAVNGLRALPTLGVLL
MMAR_0033|M.marinum_M VISLSATVLTCVVAIPVGIALTRGSMRRYSKPIITVAGFGQAAPAIGLIA
. .: . : : . :.:
Mflv_3159|M.gilvum_PYR-GCK LLIIWMGIGEGPKIVLITMGVAIAVYINTYSGIRGVDQQLVEMAQTLEAR
TH_4654|M.thermoresistible__bu LAIIWFGIGDTMKIVIIATSVLIPVYINTTAQLKGVDLRHVELAQTVGLS
MAV_2433|M.avium_104 LFILWFGIGELPKVLLVALGVVFPLYLNTFSAIRQVDPKMLETAQVLGFS
Mvan_3413|M.vanbaalenii_PYR-1 LFIIWFGIGELPKVLLVALGVSFPLYLNTFSAIRQVDPKLFETAQVLGFS
MAB_2216|M.abscessus_ATCC_1997 LFIVWFGIGELPKVLMVALGAAFPLYLNTTSAIRQVDPKLFETAQVLGFS
MSMEG_1161|M.smegmatis_MC2_155 VFVLILGIGETSKVALVVYASAFPILLNTISGVRTVDPLLIKSARSLGLS
Mb3782c|M.bovis_AF2122/97 LGVLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMT
Rv3756c|M.tuberculosis_H37Rv LGVLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMT
MUL_4374|M.ulcerans_Agy99 LGVLLFGLGLGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMT
MMAR_0033|M.marinum_M LGAVVFGIGQLGAIVALTVYGAMPIIANTVTGLDGVDRRIVEAARGMGLS
: : :*:* : : .: .* : : ** .. *: :
Mflv_3159|M.gilvum_PYR-GCK RGTLITQVILPGAMPNFLVGLRLGLSSAWLSLIFAEMINTTEGIGFLMSR
TH_4654|M.thermoresistible__bu RVQFIRKVALPGALPGFFTGLRLAVTISWTALVVVEQVNATSGVGFLMTQ
MAV_2433|M.avium_104 FFQRFRRILVPGTAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGFLINN
Mvan_3413|M.vanbaalenii_PYR-1 FWQRFRSIIVPSAAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGFLIMN
MAB_2216|M.abscessus_ATCC_1997 FWQRLRVIVIPGATPQVLVGLRQSLALSWLTLIVAEQINADAGVGFLINN
MSMEG_1161|M.smegmatis_MC2_155 PVRLFQKVVLPAAVPTIFTGLRMAAASSILVLIAAEMVGAKAGLGYLITA
Mb3782c|M.bovis_AF2122/97 ESQVLLRVEVPNALPLMLGGLR-SATLQVVATATVAAYASLGGLGGYLID
Rv3756c|M.tuberculosis_H37Rv ESQVLLRVEVPNALPLMLGGLR-SATLQVVATATVAAYASLGGLGGYLID
MUL_4374|M.ulcerans_Agy99 EAQVVLRVEMPNALPLILGGLR-SATLQVVATVTVAAYASLGGLGRYLID
MMAR_0033|M.marinum_M AFSTLRRVELPLSLPVIVAGVR-TALVLIVGTAALASFTGGGGLGQLITT
. : :* : * .. *:* *:* :
Mflv_3159|M.gilvum_PYR-GCK AQTNLQFDVSLLVIVIYAVLGLASYALVRLLERLLLSWRNGFAGFEATA-
TH_4654|M.thermoresistible__bu ARLYGQLEVVVVGLVVYAVFGLAGDHLVRTIERRALSWRRALGG------
MAV_2433|M.avium_104 ARDFLRIDIIIFGLTIYALLGIITDAVVRLIERRAVRYRH----------
Mvan_3413|M.vanbaalenii_PYR-1 ARDFLRIDIIIFGLIVYALLGIGTDAVVRALERRALRYRS----------
MAB_2216|M.abscessus_ATCC_1997 ARDFLRIDVIIFGLVVYALLGIVTDAIVRVLERRALRYRS----------
MSMEG_1161|M.smegmatis_MC2_155 SQLNFQIPNMYAGIVTIALVGVVFNSVLVAIEGRLSGWRNTN--------
Mb3782c|M.bovis_AF2122/97 GIKERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKL-----
Rv3756c|M.tuberculosis_H37Rv GIKERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKL-----
MUL_4374|M.ulcerans_Agy99 GIKERQSHIALVGALMVAALALSLDGLLALAGWASVPGTGRVSRFWHQPD
MMAR_0033|M.marinum_M GIKLQQPVTLIVGAILVAALALFIDWLARLIEMVAAPRGL----------
. : * ..: :
Mflv_3159|M.gilvum_PYR-GCK --------------------
TH_4654|M.thermoresistible__bu --------------------
MAV_2433|M.avium_104 --------------------
Mvan_3413|M.vanbaalenii_PYR-1 --------------------
MAB_2216|M.abscessus_ATCC_1997 --------------------
MSMEG_1161|M.smegmatis_MC2_155 --------------------
Mb3782c|M.bovis_AF2122/97 ----AAVVDKPAAGGGHALR
Rv3756c|M.tuberculosis_H37Rv ----AAVVDKPAAGGGHALR
MUL_4374|M.ulcerans_Agy99 DRIDVAVSANSAAVGSHVLR
MMAR_0033|M.marinum_M --------------------