For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSIIERAPRAVPVGTVAAQKALLLSRIWPPALAIAATVAVWWLASSVLSQPYSLLRQTAPDKAFRAAMDL LNRGVLLQDAGISLWRLLIGLCLAAVIGVPAGLLLGVSTTAERAARPVIQFLRMISPLSWTPIAVAVFGI GNQPVVFLIATAAVWPILLNTTAGVHSIEPGYLHVARSFHATRSEVLTAVILPAIRGHIQTGIRVALGVA WVVLVPAEMLGVRSGLGYQVLNARDQLAYDQVVAVILVIGILGYALDMAAKRLLASRFRVPSAG
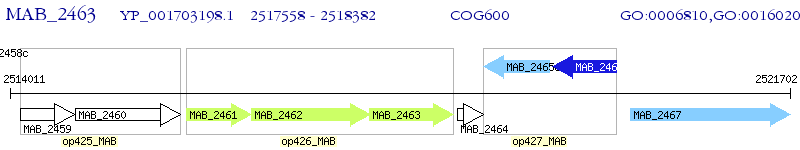
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2463 | - | - | 100% (274) | putative sulfonate ABC transporter, permease |
| M. abscessus ATCC 19977 | MAB_2216 | - | 3e-28 | 29.64% (253) | sulfonate ABC transporter, permease protein SsuC |
| M. abscessus ATCC 19977 | MAB_2242 | - | 2e-24 | 30.13% (239) | putative nitrate ABC transporter, permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3782c | proZ | 2e-09 | 25.53% (188) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3159 | - | 2e-26 | 29.70% (266) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3756c | proZ | 2e-09 | 25.53% (188) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5299 | proZ | 1e-09 | 23.94% (188) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. avium 104 | MAV_2433 | - | 4e-30 | 28.95% (266) | putative aliphatic sulfonates transport permease protein SsuC |
| M. smegmatis MC2 155 | MSMEG_4523 | - | 1e-93 | 68.92% (251) | binding-protein-dependent transport systems inner membrane |
| M. thermoresistible (build 8) | TH_4654 | - | 6e-23 | 29.79% (235) | PUTATIVE putative binding-protein-dependent transport systems |
| M. ulcerans Agy99 | MUL_4374 | proZ | 3e-10 | 26.09% (184) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_3413 | - | 2e-29 | 30.09% (226) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2463|M.abscessus_ATCC_1997 MSIIERAPRAVP-----------VGTVAAQKAL-------LLSRIWPPAL
MSMEG_4523|M.smegmatis_MC2_155 MTIAGTRNGPTT-----------AGEVSWQR------------RCAPPIT
Mflv_3159|M.gilvum_PYR-GCK MTAVAAPLTTP--RGPAAPAPELVDVIAGHRGR-RSIWG-RIPRPVRRMA
TH_4654|M.thermoresistible__bu VTSLQTARPAPRLRSPAQRRADVTDAQLLHRGEHRTRLGPGRAVPAALAV
MAV_2433|M.avium_104 MSIATALDAAP---------KRLGGAASGAPGG----LWARRKWEVVRLV
Mvan_3413|M.vanbaalenii_PYR-1 MTRPADPTAP------------LAQANSGRVG-----RRADRRWALIRLA
Mb3782c|M.bovis_AF2122/97 --------------------------------------------------
Rv3756c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5299|M.marinum_M --------------------------------------------------
MUL_4374|M.ulcerans_Agy99 --------------------------------------------------
MAB_2463|M.abscessus_ATCC_1997 AIAATVAVWWLASSVLSQPYSLLRQTAPDKAFRAAMDLLNRGVLLQDAGI
MSMEG_4523|M.smegmatis_MC2_155 AILIAIALWWSATSVLSGPESVLRQTAPQHVVPALSGLLERQVLLGDLGV
Mflv_3159|M.gilvum_PYR-GCK GPVAILAVWTIGSTSGLLNTDLFPP--PSDVAVTAWHLLTNGGLALHVGT
TH_4654|M.thermoresistible__bu GPLLLVALWLITSQLGILDPRTLPH--PFDIARTAAELWSDGRLPANVAA
MAV_2433|M.avium_104 SPLALLALWQLGSAIGVIAQDVLPA--PSLILQAGVELTRNGQLADALHI
Mvan_3413|M.vanbaalenii_PYR-1 SPFILLAVWQLGSAVGLIPQDVLPA--PSLIAEAGIELIGNGQLVDALTV
Mb3782c|M.bovis_AF2122/97 -------------------MNFLQQ--ALSYLLTASNWTGPVGLAVRTCE
Rv3756c|M.tuberculosis_H37Rv -------------------MNFLQQ--ALSYLLTASNWTGPVGLAVRTCE
MMAR_5299|M.marinum_M -------------------MNFLQQ--ALSYLSTAANWTGSAGLAHRTAE
MUL_4374|M.ulcerans_Agy99 -------------------MNFLQQ--ALSYLSTAANWTGSAGLAHRTAE
: . : *
MAB_2463|M.abscessus_ATCC_1997 SLWRLLIGLCLAAVIGVPAGLLLGVSTTAERAARPVIQFLRMISPLSWTP
MSMEG_4523|M.smegmatis_MC2_155 SLWRLSIGLAVATVIGMPLGLLIGSSRIAERAGGPIIAFLRMISPLSWTP
Mflv_3159|M.gilvum_PYR-GCK SATRVLIGTALGITVGVVLAVLAGLTRTGEDLLDWSMQILKAVPNFALTP
TH_4654|M.thermoresistible__bu SLKLSLISLAIGVAAGLALALLAGLSRIGEALVDGPIQIKRAVPTLAIIP
MAV_2433|M.avium_104 STVRVIEGLALGGVIGIVAGAAVGLSRWVEATVDPPLQMIRALPHLGLIP
Mvan_3413|M.vanbaalenii_PYR-1 SGLRVVEGLILGGVIGIALGTAVGLSKWFEATVDPPMQMVRALPHLGLIP
Mb3782c|M.bovis_AF2122/97 HLEYTAVAVAASALIAVPVGLLIGHTGRGTLLVVGAVNGLRALPTLGVLL
Rv3756c|M.tuberculosis_H37Rv HLEYTAVAVAASALIAVPVGLLIGHTGRGTLLVVGAVNGLRALPTLGVLL
MMAR_5299|M.marinum_M HLEYTALAVTASALVAIPIGLIIGHTGRGQLLVVSAVNGLRALPTLGVLL
MUL_4374|M.ulcerans_Agy99 HLEYTALAATASALVAIPIGLIIGHTGRGQLLVVSAVNGLRALPTLGVLL
. . .: . * : : : :. :.
MAB_2463|M.abscessus_ATCC_1997 IAVAVFGIGNQPVVFLIATAAVWPILLNTTAGVHSIEPGYLHVARSFHAT
MSMEG_4523|M.smegmatis_MC2_155 IALAVFGIGNQPVVFLVAVAAVWPVLLNTAAGVRAVDPGLLNVARSFHAT
Mflv_3159|M.gilvum_PYR-GCK LLIIWMGIGEGPKIVLITMGVAIAVYINTYSGIRGVDQQLVEMAQTLEAR
TH_4654|M.thermoresistible__bu LAIIWFGIGDTMKIVIIATSVLIPVYINTTAQLKGVDLRHVELAQTVGLS
MAV_2433|M.avium_104 LFILWFGIGELPKVLLVALGVVFPLYLNTFSAIRQVDPKMLETAQVLGFS
Mvan_3413|M.vanbaalenii_PYR-1 LFIIWFGIGELPKVLLVALGVSFPLYLNTFSAIRQVDPKLFETAQVLGFS
Mb3782c|M.bovis_AF2122/97 LGVLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMT
Rv3756c|M.tuberculosis_H37Rv LGVLLFGLGLGPPLVALMLLGIPSLLASTYAGIASVDPLVVDAARAMGMT
MMAR_5299|M.marinum_M LGVLLFGLGLGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMT
MUL_4374|M.ulcerans_Agy99 LGVLLFGLGLGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMT
: : :*:* :. : .: .* : : :: .. *: .
MAB_2463|M.abscessus_ATCC_1997 RSEVLTAVILPAIRGHIQTGIRVALGVAWVVLVPAEMLGVRSGLGYQVLN
MSMEG_4523|M.smegmatis_MC2_155 RWEMMLAVILPAIRTHVQTGIRLALGVAWIVLVPAEMLGVRSGLGYQILN
Mflv_3159|M.gilvum_PYR-GCK RGTLITQVILPGAMPNFLVGLRLGLSSAWLSLIFAEMINTTEGIGFLMSR
TH_4654|M.thermoresistible__bu RVQFIRKVALPGALPGFFTGLRLAVTISWTALVVVEQVNATSGVGFLMTQ
MAV_2433|M.avium_104 FFQRFRRILVPGTAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGFLINN
Mvan_3413|M.vanbaalenii_PYR-1 FWQRFRSIIVPSAAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGFLIMN
Mb3782c|M.bovis_AF2122/97 ESQVLLRVEVPNALPLMLGGLR-SATLQVVATATVAAYASLGGLGGYLID
Rv3756c|M.tuberculosis_H37Rv ESQVLLRVEVPNALPLMLGGLR-SATLQVVATATVAAYASLGGLGGYLID
MMAR_5299|M.marinum_M EAQVVLRVEMPNALPLILGGLR-SATLQVVATATVAAYASLGGLGRYLID
MUL_4374|M.ulcerans_Agy99 EAQVVLRVEMPNALPLILGGLR-SATLQVVATVTVAAYASLGGLGRYLID
. : :* . *:* . . *:* :
MAB_2463|M.abscessus_ATCC_1997 ARDQLAYDQVVAVILVIGILGYALDMAAKRLLASRFRVPSAG--------
MSMEG_4523|M.smegmatis_MC2_155 ARDQLAYDQVVAVIVVIGILGFLLDTTARWLLNPARRA------------
Mflv_3159|M.gilvum_PYR-GCK AQTNLQFDVSLLVIVIYAVLGLASYALVRLLERLLLSWRNGFAGFEATA-
TH_4654|M.thermoresistible__bu ARLYGQLEVVVVGLVVYAVFGLAGDHLVRTIERRALSWRRALGG------
MAV_2433|M.avium_104 ARDFLRIDIIIFGLTIYALLGIITDAVVRLIERRAVRYRH----------
Mvan_3413|M.vanbaalenii_PYR-1 ARDFLRIDIIIFGLIVYALLGIGTDAVVRALERRALRYRS----------
Mb3782c|M.bovis_AF2122/97 GIKERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKL-----
Rv3756c|M.tuberculosis_H37Rv GIKERRFHIALVGAMMVAALALTLDGLLALAGWVSVPGTGRMRKL-----
MMAR_5299|M.marinum_M GIKERQFHIALVGALMVAALALSLDGLLALAGWASVPGTGRLSRFWHQSD
MUL_4374|M.ulcerans_Agy99 GIKERQSHIALVGALMVAALALSLDGLLALAGWASVPGTGRVSRFWHQPD
. . : : . :.
MAB_2463|M.abscessus_ATCC_1997 --------------------
MSMEG_4523|M.smegmatis_MC2_155 --------------------
Mflv_3159|M.gilvum_PYR-GCK --------------------
TH_4654|M.thermoresistible__bu --------------------
MAV_2433|M.avium_104 --------------------
Mvan_3413|M.vanbaalenii_PYR-1 --------------------
Mb3782c|M.bovis_AF2122/97 ----AAVVDKPAAGGGHALR
Rv3756c|M.tuberculosis_H37Rv ----AAVVDKPAAGGGHALR
MMAR_5299|M.marinum_M DRIDVAVSPNSAAVGSHVLR
MUL_4374|M.ulcerans_Agy99 DRIDVAVSANSAAVGSHVLR