For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIGELAERTGVSRRQLRYYEEQGLLMPNRAANGYREYGEAHVHVVLQIAGLLDAGLPTRIIEQLLPCLDQ PQTIYVPNVTPEMVATLQREQARLTERIECLARNRDAIAAYLDTVLSVRSANLARERQGS*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
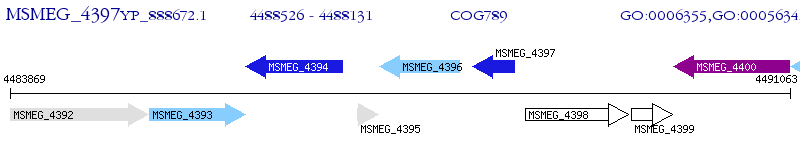
| M. smegmatis MC2 155 | MSMEG_4397 | - | - | 100% (131) | transcriptional regulator, MerR family protein |
| M. smegmatis MC2 155 | MSMEG_1969 | - | 6e-15 | 41.28% (109) | MerR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_4765 | - | 1e-06 | 35.14% (74) | transcriptional regulator, MerR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3367 | - | 1e-06 | 40.28% (72) | MerR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3212 | - | 2e-07 | 33.94% (109) | MerR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3334 | - | 1e-06 | 40.28% (72) | MerR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2246 | - | 7e-14 | 35.96% (114) | thiol-specific antioxidant related protein/peroxidoxin BcpB |
| M. marinum M | MMAR_0009 | - | 1e-07 | 33.67% (98) | transcriptional regulator |
| M. avium 104 | MAV_4105 | - | 2e-06 | 28.42% (95) | transcriptional regulator, MerR family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0011 | - | 9e-08 | 33.67% (98) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1919 | - | 9e-06 | 44.00% (50) | MerR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4397|M.smegmatis_MC2_155 ------------------MRIGELAERTGVSRRQLRYYEEQGLLMPN-RA
MAB_2246|M.abscessus_ATCC_1997 ------------------MQISELARRAGVSLKAVRYYERLGLVMPS-RL
Mb3367|M.bovis_AF2122/97 ------------------MKISEVAALTNTSTKTLRFYENSGLLPPPART
Rv3334|M.tuberculosis_H37Rv ------------------MKISEVAALTNTSTKTLRFYENSGLLPPPART
Mflv_3212|M.gilvum_PYR-GCK ------------------MRIGKLAEATGATTATLRYYEDEGLLPPAERS
MMAR_0009|M.marinum_M ---------------MDGLTVGQVAERFGITVRTLHHYDEIGLLIPSRRA
MUL_0011|M.ulcerans_Agy99 ---------------MDGLTVGQVAERFGITVRTLHHYDEIGLLIPSRRA
Mvan_1919|M.vanbaalenii_PYR-1 MTSGTTLTYVTQEMQVDELTVGEVAERFGITVRTLHHYDEIGLLTPGRRC
MAV_4105|M.avium_104 -MADRRGNAGAPASDQGVYGISVAAELSGIAVQSLRLYERHGLVTPA-RS
:. * . : :: *: **: * *
MSMEG_4397|M.smegmatis_MC2_155 ANGYREYGEAHVHVVLQIAGLLDAGLPTRIIEQLLPCLDQPQTIYVPNVT
MAB_2246|M.abscessus_ATCC_1997 GNGYRDYTDDDLQIVGEIRSLAAAGISPSLARPFIECLREG--HDHSDDC
Mb3367|M.bovis_AF2122/97 ASGYRNYGPEIVDRLRFIHRGQAAGLALQEVRQILAIHDRG-----EAPC
Rv3334|M.tuberculosis_H37Rv ASGYRNYGPEIVDRLRFIHRGQAAGLALQEVRQILAIHDRG-----EAPC
Mflv_3212|M.gilvum_PYR-GCK PAGYRDYAADTIARVGFIRRGQAAGFSLAQIRQILDIRDSG-----HAPC
MMAR_0009|M.marinum_M ASGYRVYTSADLTRLSQVIVYRRLELPLDEIARLLDE---------GNAV
MUL_0011|M.ulcerans_Agy99 ASGYRVYTSADLTRLSQVIVYRRLELPLDEIARLLDE---------GNAV
Mvan_1919|M.vanbaalenii_PYR-1 ASGYRVYTPADLTRLSQVIVYRRLDFSLDEIASLLDE---------GNEV
MAV_4105|M.avium_104 QGGTRRYSADDLARLKRISKLVDAGVNLAGVARILALEDDN---------
* * * : : : . ::
MSMEG_4397|M.smegmatis_MC2_155 PEMVATLQREQARLTERIECLARNRDAIAAYLDTVLSVRSANLARERQGS
MAB_2246|M.abscessus_ATCC_1997 PSALAAYRDSIAALDQSIASLIMRRRILAARLDAAAARTFTEEPRNMDDY
Mb3367|M.bovis_AF2122/97 AHVRQLLSTRIDEVRAQIAELIALEGHLQTLLDHASYGPPTEHDHSTVCW
Rv3334|M.tuberculosis_H37Rv AHVRQLLSTRIDEVRAQIAELIALEGHLQTLLDHASYGPPTEHDHSTVCW
Mflv_3212|M.gilvum_PYR-GCK THVRDLLDIRLTDLDEQISALVALRETIARLRQGAESVDPESCSADDVCR
MMAR_0009|M.marinum_M SHLVRQRERVMARLDEMKDLVEAIDQALEKAMTNTPMTDDDMRELFGDGF
MUL_0011|M.ulcerans_Agy99 SHLVRQRERVMARLDEMKDLVEAIDLALEKAMTNTPMTDDDMRELFGDGF
Mvan_1919|M.vanbaalenii_PYR-1 SHLIRQRERVMSRLDEMKDLVAAIDQALEKAMTNTPMTDDDMRELFGDGF
MAV_4105|M.avium_104 -----------ATLSAANTDLRSTNRALREAAKTADGDAATG--------
: : : .
MSMEG_4397|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2246|M.abscessus_ATCC_1997 TELPFGLPVPVDDGAADHLQDRPLPDLTLMATGGCAVRLRGLPEGRTVLY
Mb3367|M.bovis_AF2122/97 ILESDLDEPTAIEVSDIHA-------------------------------
Rv3334|M.tuberculosis_H37Rv ILESDLDEPTAIEVSDIHA-------------------------------
Mflv_3212|M.gilvum_PYR-GCK YL------------------------------------------------
MMAR_0009|M.marinum_M DDYRAETEQKWGETTEWKESQRR------TRSYGKDEWVRIKAEGQAVEK
MUL_0011|M.ulcerans_Agy99 DDYRVETEQKWGETAEWKESQRR------TKSYGKDEWVRIKAEGQAIEK
Mvan_1919|M.vanbaalenii_PYR-1 DDYQAEAEQKWGETAEWKESQRR------TKSYGKDEWVQIKSEGEAVEK
MAV_4105|M.avium_104 --------------------------------------------------
MSMEG_4397|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2246|M.abscessus_ATCC_1997 LYPLTGRPGLDIPEGWDAIPGARGCTTEACGFRDHFDELRAAGAQQVWGL
Mb3367|M.bovis_AF2122/97 --------------------------------------------------
Rv3334|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3212|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0009|M.marinum_M TLSDAFRAGLPADS----------------------EEAMNAAEQHRRHV
MUL_0011|M.ulcerans_Agy99 ALSDAFRAGLPADS----------------------EEAMNAAEQHRRHV
Mvan_1919|M.vanbaalenii_PYR-1 ALSDAFRAGLPPDS----------------------AEAMNAAEQHRLHV
MAV_4105|M.avium_104 --------------------------------------------------
MSMEG_4397|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2246|M.abscessus_ATCC_1997 SSQDTDYQAEVVRRLRLPFHMLSDPDFTLAEALHLPTFAAVGHPRLYARI
Mb3367|M.bovis_AF2122/97 --------------------------------------------------
Rv3334|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3212|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0009|M.marinum_M NRWFYDCPPAFHRSLGDMYVSDPRYIATYDESFGLPGLAAYCREAIHANA
MUL_0011|M.ulcerans_Agy99 NRWFYDCPPAFHRNLGDMYVSDPRYIATYDESFGLPGLAAYCREAIHANA
Mvan_1919|M.vanbaalenii_PYR-1 NRWFYDCPPAFHRKLGDMYVSDPRYVATYDESFGLPGLAAYCREAIHANA
MAV_4105|M.avium_104 --------------------------------------------------
MSMEG_4397|M.smegmatis_MC2_155 ------------------------------------
MAB_2246|M.abscessus_ATCC_1997 TLVVRDGRIEQVFYPIFPPDTHAQQVLEWLTSHPMD
Mb3367|M.bovis_AF2122/97 ------------------------------------
Rv3334|M.tuberculosis_H37Rv ------------------------------------
Mflv_3212|M.gilvum_PYR-GCK ------------------------------------
MMAR_0009|M.marinum_M NRTGG-------------------------------
MUL_0011|M.ulcerans_Agy99 NRAGG-------------------------------
Mvan_1919|M.vanbaalenii_PYR-1 DRAGQ-------------------------------
MAV_4105|M.avium_104 ------------------------------------