For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVREAAGTETMRAIVLTGFGGLDKLVYTEIPKPRPKHGEVVIAVKAFGLNHAEMHMRRGEWAEAAEVSGI ECVGVVDSCPDGTFDVGDRVAALMGGLGRTINGSYAEYTRVRTANVARIDVDLPWSQLAALPETYATAWT CLFRSLELEPGQLLLIRGATSSFGQAALKMAVNAGARVIATTRKPEQFTTLTAMGALRGELEGPDLARRL PEAKSVDAVLDLVGNSTILDSLDLLHRGGRACLAGWLGGLAPIPDFNPLARMASGVYLTFFGSFVFGTPG FPLADVPLSDIAAQVQAGEFDASPVRVFSFDDIREAHQVMESGHANGKMVVTLD
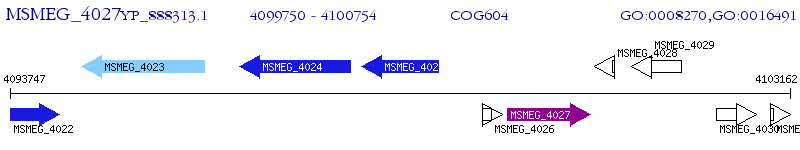
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4027 | - | - | 100% (334) | zinc-containing alcohol dehydrogenase superfamily protein |
| M. smegmatis MC2 155 | MSMEG_4067 | - | 1e-26 | 30.06% (336) | zinc-containing alcohol dehydrogenase superfamily protein |
| M. smegmatis MC2 155 | MSMEG_6362 | - | 7e-26 | 27.58% (330) | quinone oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3806 | - | 3e-26 | 29.34% (334) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1189 | - | 4e-27 | 28.00% (325) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3777 | - | 1e-25 | 29.04% (334) | oxidoreductase |
| M. leprae Br4923 | MLBr_00118 | - | 7e-24 | 26.25% (320) | putative oxidireductase |
| M. abscessus ATCC 19977 | MAB_0207c | - | 2e-26 | 27.03% (333) | quinone oxidoreductase |
| M. marinum M | MMAR_5332 | - | 3e-25 | 27.24% (323) | oxidoreductase |
| M. avium 104 | MAV_0242 | - | 4e-24 | 28.53% (333) | quinone oxidoreductase |
| M. thermoresistible (build 8) | TH_2952 | - | 2e-25 | 30.09% (329) | PUTATIVE Alcohol dehydrogenase, zinc-binding domain protein |
| M. ulcerans Agy99 | MUL_0096 | - | 5e-25 | 26.93% (323) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5758 | - | 2e-23 | 30.84% (334) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00118|M.leprae_Br4923 ----------MHAIVAESA---NQMVWREVPDVTAGPGEVLIEVAASGVN
MAV_0242|M.avium_104 -------MATMRAIVAESA---DQLSWQEVADVSAGPGEVVVKVAAAGVN
Mb3806|M.bovis_AF2122/97 -------MTIMRAVVAESS---DRLVWQEVPDVSAGPGEVLIKVAASGVN
Rv3777|M.tuberculosis_H37Rv -------MTIMRAVVAESS---DRLVWQEVPDVSAGPGEVLIKVAASGVN
MMAR_5332|M.marinum_M -------MAFVHAIVAESA---DHLVWQQVPDVTAGAGEVVIKVAAAGVN
MUL_0096|M.ulcerans_Agy99 -------MAFVHAIVAESA---DHLVWQQVPDVTAGAGEVVIKVAAAGVN
Mflv_1189|M.gilvum_PYR-GCK --------------MAETP---DQLNWQSVPDPTPGPGEVLIRVHAAGVN
MAB_0207c|M.abscessus_ATCC_199 ----------MHAITVDNPGGPEALRWTEVPDPVPGPGEILIDVTAAAAN
TH_2952|M.thermoresistible__bu ----------VKALVAQELSGPSGLVYTDWEDPSENDDVVVIDVRAAGLS
MSMEG_4027|M.smegmatis_MC2_155 MVREAAGTETMRAIVLTGFGGLDKLVYTEIPKPRPKHGEVVIAVKAFGLN
Mvan_5758|M.vanbaalenii_PYR-1 ----------MRAAVLTAYN--SPLLVDELPDPEPGPGEVLVRVMASGVN
. : . . . ::: * * . .
MLBr_00118|M.leprae_Br4923 RADVLQVAG-KYPHPPGASAIIGMEVAGVVAAVSSGVTEWSVGQEVCALL
MAV_0242|M.avium_104 RADVLQAAG-KYPPPPGASETIGMEVSGVIAEVGDGVTEWSVGQEVCALL
Mb3806|M.bovis_AF2122/97 RADVLQAAG-KYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALL
Rv3777|M.tuberculosis_H37Rv RADVLQAAG-KYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALL
MMAR_5332|M.marinum_M RADVLQAAG-KYPPPPGASEIIGMEVSGTITDIGDGVTEWSAGQEVCALL
MUL_0096|M.ulcerans_Agy99 RADVLQAAG-KYPPPPGASEIIGMEVSGTITDIGDGVTEWSAGQEVCALL
Mflv_1189|M.gilvum_PYR-GCK RADLLQAAG-KYPPPPGASEIIGLEASGTIAAVGDGVTEWSEGQRVCALL
MAB_0207c|M.abscessus_ATCC_199 RADLLQRRG-LYSPPPGASSILGLECSGTVAALGADVAHWSVGQPVCALL
TH_2952|M.thermoresistible__bu FPDLLLIRG-EYQLRLDPPFIPGMEIAGEVRSAPQGS-GFRPGQRVSAFS
MSMEG_4027|M.smegmatis_MC2_155 HAEMHMRRG-EWA---EAAEVSGIECVGVVDSCPDGT--FDVGDRVAALM
Mvan_5758|M.vanbaalenii_PYR-1 PLDIKIRRGEAPHAQLPPPAVLGIDMAGVVEAVGPRVDRFTVGDEVFGMT
:: * . *:: * : : *: * .:
MLBr_00118|M.leprae_Br4923 A------GGGYAEYVAVPASQVMPIPKGVNVVDSAGIPEVACTVWSNLVM
MAV_0242|M.avium_104 A------GGGYAEYVAVPAGQLLPIPAGVDLVDAAGLPEVACTVWSNLVL
Mb3806|M.bovis_AF2122/97 A------GGGYAEYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVM
Rv3777|M.tuberculosis_H37Rv A------GGGYAEYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVM
MMAR_5332|M.marinum_M A------GGGYAEYVAVPAGQVLPMPPGVSLIDAAALPEVACTVWSNLVM
MUL_0096|M.ulcerans_Agy99 A------GGGYAEYVAVPAGQVLPMPPGVSLIDAAALPEVACTVWSNLVM
Mflv_1189|M.gilvum_PYR-GCK A------GGGYAEYVAVPAGQVLPVPDGVPLDHAAGLPEVACTVWSNLVM
MAB_0207c|M.abscessus_ATCC_199 S------GGGYAEKVVAPATQLLPVPTNVQLDVAAALPEVACTVWSNMMM
TH_2952|M.thermoresistible__bu M------MGGFAERVAVAPQNVIPTADGIDDASAAALLGNYYTMYFALAR
MSMEG_4027|M.smegmatis_MC2_155 GGLGRTINGSYAEYTRVRTANVARIDVDLPWSQLAALPETYATAWTCLFR
Mvan_5758|M.vanbaalenii_PYR-1 GGVGG-LQGSLAELAAVDARLIAHKPRTLSMAQAAALPLGFITSWEGLVD
*. ** . . . : : *.: * : :
MLBr_00118|M.leprae_Br4923 AAHLSAGQLLLLHGGTSGIGSHGIQVARALGAKVAVTAGSEEKLEFCRAL
MAV_0242|M.avium_104 IAGLRNGQLLLVHGGASGIGSHAIQVARALGARVAVTAGSAEKLGFCREL
Mb3806|M.bovis_AF2122/97 TAHLRPGQLVLIHGGASGIGSHAIQVARALAARVAITAGSPEKLELCRDL
Rv3777|M.tuberculosis_H37Rv TAHLRPGQLVLIHGGASGIGSHAIQVVRALAARVAITAGSPEKLELCRDL
MMAR_5332|M.marinum_M TAHLGPGQLVLIHGGASGIGSHAIQVCTALGARVAVTAGSSEKLQLCRDL
MUL_0096|M.ulcerans_Agy99 TAHLGPGQLVLIHGGACGIGSHAIQVCTALGARVAVTAGSSEKLQLSRDL
Mflv_1189|M.gilvum_PYR-GCK TARLRAPELLLIHGGASGIGTHAIQVARALNCRVAVTAGSRNKLDLCAEL
MAB_0207c|M.abscessus_ATCC_199 TTKMSAGQSILIHGGASGIGTHTIQVAKALGLRVAVTAGTPEKLDACRDL
TH_2952|M.thermoresistible__bu RAALRPGETVLVFGSAGGVGTAAIQIAKAMGARVIAMVRRARGTEYVESL
MSMEG_4027|M.smegmatis_MC2_155 SLELEPGQLLLIRGATSSFGQAALKMAVNAGARVIATTRKPEQFTTLTAM
Mvan_5758|M.vanbaalenii_PYR-1 RAGVGTGQQVLIHGGAGGVGFLAVQLALARGAQVYATGSTSS--QAVIRA
: : :*: *.: ..* ::: :*
MLBr_00118|M.leprae_Br4923 GAQITINYRDEDFVARLQ-QQN--GGADVILDIMGAAYLDRNIDALAIDG
MAV_0242|M.avium_104 GADITINYRDEDFVARLA-DET--GGADVILDIMGAAYLDRNIDALATDG
Mb3806|M.bovis_AF2122/97 GAQITINYRDEDFVARLK-QETDGSGADIILDIMGASYLDRNIDALATDG
Rv3777|M.tuberculosis_H37Rv GAQITINYRDEDFVARLK-QETDGSGADIILDIMGASYLDRNIDALATDG
MMAR_5332|M.marinum_M GAQITINYRDEDFVARLK-QETDGSGADVILDIMGAAYLDRNVDALAVDG
MUL_0096|M.ulcerans_Agy99 GAQITINYRDEDFVARLK-QETDGSGADVILDIMGAAYLDRNVDALAVDG
Mflv_1189|M.gilvum_PYR-GCK GAEITIDYHHEDFVEVLRGADSDHPGADVILDIMGASYLDRNIDALATGG
MAB_0207c|M.abscessus_ATCC_199 GADLAINYRDTDFVQAVH-SFTDGSGVDRILDIMGAKYLSRNVDALGNDG
TH_2952|M.thermoresistible__bu GPDVVLPLTEG-WPQAVR-DATDGRGVDVVVDPVGGDVFDDAIRVLATEG
MSMEG_4027|M.smegmatis_MC2_155 G-ALRGELEGPDLARRLP----EAKSVDAVLDLVGNSTILDSLDLLHRGG
Mvan_5758|M.vanbaalenii_PYR-1 AGAVPIDYTATTVDEYVA-AHTGGEGFDVVADNVGGATLDASFTAVKRYT
. : : . * : * :* : . :
MLBr_00118|M.leprae_Br4923 QLIVIGLQGG-VKGELNLGKVLSKRARIIGSTLRARPVNGPNSKSEIVAA
MAV_0242|M.avium_104 QLVIIGMQGG-VKAELNIGKLLAKRARVIGTTLRGRPVSGPNSKTEIVQA
Mb3806|M.bovis_AF2122/97 QLIVIGMQGG-VKAELNLGKLLTKRARVIGTTLRARPVSGPHGKAAIAQA
Rv3777|M.tuberculosis_H37Rv QLIVIGMQGG-VKAELNLGKLLTKRARVIGTTLRARPVSGPHGKAAIAQA
MMAR_5332|M.marinum_M QLVIIGMQGG-IKAELNLGKLLTKRARVIGTTLRARPLQGPNGKAVIAQA
MUL_0096|M.ulcerans_Agy99 QLVIIGMQGG-IKAELNLGKLLTKRARVIGTTLRARPLQGPNGKAVIAQA
Mflv_1189|M.gilvum_PYR-GCK RLVIIGMQGG-TKAEINLGKLLAKRGSVMATALRSRPVTGRGSKSVIVSE
MAB_0207c|M.abscessus_ATCC_199 HLVIIGMQGG-VTAELNIAKLIAKRGSLTATTLRARPIAGPCGKAAIVES
TH_2952|M.thermoresistible__bu RLLVLGFAAGQGIPTVKVNRLLLRNVGVIGVGYGEFVARNPG-----AQQ
MSMEG_4027|M.smegmatis_MC2_155 RACLAGWLGG-LAPIPDFNPLARMASGVYLTFFGSFVFGTPG--FPLADV
Mvan_5758|M.vanbaalenii_PYR-1 GHVVSALGWG----THSLAPLSFRGAAYSGVFTLMPLLTGRG--RDHHGE
: . * .. :
MLBr_00118|M.leprae_Br4923 VTASIWPMIADGSVRP-IIGARMAVQQAADAHQLLLSGKVSGKIVLTVAG
MAV_0242|M.avium_104 VTASVWPMIADGRVRP-IIGARLPIQQAGDAHRRLVASEVTGKIVLTV--
Mb3806|M.bovis_AF2122/97 VAASVWPMIAANRVRP-VIGTRLPIQQAAQAHELMLSGKTFGKILLTV--
Rv3777|M.tuberculosis_H37Rv VAASVWPMIAANRVRP-VIGTRLPIQQAAQAHELMLSGKTFGKILLTV--
MMAR_5332|M.marinum_M VTASVWPMIASNRVRP-IIGSRLPIGQAAQAHQLLLSGQSYGKTLLTVDG
MUL_0096|M.ulcerans_Agy99 VTASVWPMIASNRVRP-IIGSRLPIGQAAQAHQLLLSGQSYGKILLTVDG
Mflv_1189|M.gilvum_PYR-GCK VIENVWPLVADGQVRP-VIGAEFPISEAQAAHELLASGDVSGKVILRVDV
MAB_0207c|M.abscessus_ATCC_199 VAQNLWPLVEAGKVRP-VVGAEIPLSQAAEAHRLLENGAVVGKVLLTR--
TH_2952|M.thermoresistible__bu LFGSGLRHLVEGGLRP-PPPVRFPLSEGRAAFEALAAGEVLGKAVLEP--
MSMEG_4027|M.smegmatis_MC2_155 PLSDIAAQVQAGEFDA-SPVRVFSFDDIREAHQVMESGHANGKMVVTLD-
Mvan_5758|M.vanbaalenii_PYR-1 IVAEAAALADSGRLAPRLHPTTFPLDAVNDAHTAVESGSATGKVVVEPNV
. . . . :.. *. : . ** ::
MLBr_00118|M.leprae_Br4923 SGTETLLSRHH
MAV_0242|M.avium_104 -----------
Mb3806|M.bovis_AF2122/97 -----------
Rv3777|M.tuberculosis_H37Rv -----------
MMAR_5332|M.marinum_M -----------
MUL_0096|M.ulcerans_Agy99 -----------
Mflv_1189|M.gilvum_PYR-GCK -----------
MAB_0207c|M.abscessus_ATCC_199 -----------
TH_2952|M.thermoresistible__bu -----------
MSMEG_4027|M.smegmatis_MC2_155 -----------
Mvan_5758|M.vanbaalenii_PYR-1 -----------