For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMTQGRARVAVVTGAARGIGEATARQLHADGFAVALMDLAPEVGDLARELSGGTGNAIGIRGDVADPRAW ADAASAARTLGSVSVLVSNAMTVDIASLDETSTASWHRQLDVMLTGTFHGVKTFLPDLRAARGSIVLVSS VHAMFGLPGRPAYAAAKAGLTGLGRQLAAEYGPQLRVNTVLPGPILTASWDGIGEQDRRTSAAATALGRL GTPREVASVISFLASEASSYITGASIPVDGGWSVTKDSS
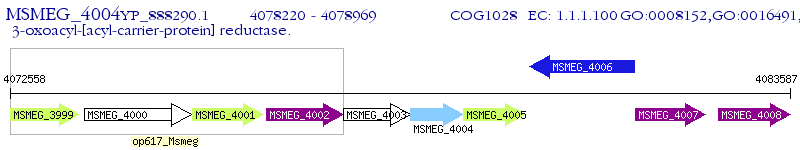
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4004 | - | - | 100% (249) | 3-oxoacyl- |
| M. smegmatis MC2 155 | MSMEG_2566 | - | 2e-34 | 40.08% (237) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0527 | - | 4e-32 | 37.30% (244) | 2-hydroxycyclohexnecarboxyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 9e-34 | 37.39% (238) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2469 | - | 5e-37 | 40.74% (243) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 9e-34 | 37.39% (238) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-30 | 37.82% (238) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0952 | - | 1e-31 | 37.25% (247) | short chain dehydrogenase |
| M. marinum M | MMAR_0402 | - | 6e-34 | 39.58% (240) | short chain dehydrogenase |
| M. avium 104 | MAV_2509 | - | 8e-34 | 36.29% (237) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_3912 | - | 9e-34 | 39.50% (238) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1052 | - | 8e-33 | 39.17% (240) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4185 | - | 1e-37 | 41.87% (246) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2469|M.gilvum_PYR-GCK ----------------MSSRVAVVTGGASGMG------ESTCHELGRRGH
Mvan_4185|M.vanbaalenii_PYR-1 -----------------MTRVAVVTGGASGMG------EATCHELGRRGC
MLBr_01807|M.leprae_Br4923 -----------------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGI
MAB_0952|M.abscessus_ATCC_1997 MVVESAEPAERKVMFDLSARSVLVTGGTKGIG------RGIATVFARAGA
MSMEG_4004|M.smegmatis_MC2_155 ------------MMTQGRARVAVVTGAARGIG------EATARQLHADGF
Mb2025|M.bovis_AF2122/97 ------------MSGRLIGKVALVSGGARGMG------ASHVRAMVAEGA
Rv2002|M.tuberculosis_H37Rv ------------MSGRLIGKVALVSGGARGMG------ASHVRAMVAEGA
MAV_2509|M.avium_104 ------------MAERLAGKVALVSGGARGMG------ASHVRSLVAEGA
MMAR_0402|M.marinum_M ------------MTG--GGRVALVTGAAGGQG------WAIVQRLRAEGH
MUL_1052|M.ulcerans_Agy99 ------------MTG--GGRVALVTGAAGGQG------WAIVQRLRAEGH
TH_3912|M.thermoresistible__bu -----------------------VTGAARGQG------SAIVRRLLQDGY
..: * * . *
Mflv_2469|M.gilvum_PYR-GCK KVAVLDLNGEAAQRVAEELRADGVT--ALGIAADVTDRAAVEDAFAKVRT
Mvan_4185|M.vanbaalenii_PYR-1 KVAVLDLDGQAAQRVAEELRGDGVP--ALGVAADVTDRAAVEDAFAKVRT
MLBr_01807|M.leprae_Br4923 GLAVARRLVADGHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAVEE
MAB_0952|M.abscessus_ATCC_1997 NVAVAARSPRELSSVTAELGELGAGN-VIGVRLDVSDPGSCADAARTVVD
MSMEG_4004|M.smegmatis_MC2_155 AVALMDLAP-EVGDLARELSG-GTGN-AIGIRGDVADPRAWADAASAAR-
Mb2025|M.bovis_AF2122/97 KVVFGDILDEEGKAVAAELA-DAARY----VHLDVTQPAQWTAAVDTAVT
Rv2002|M.tuberculosis_H37Rv KVVFGDILDEEGKAVAAELA-DAARY----VHLDVTQPAQWTAAVDTAVT
MAV_2509|M.avium_104 KVVFGDILDDEGKAVAAEVG-EATRY----LHLDVTKPEDWDAAVATALA
MMAR_0402|M.marinum_M SVAACDLRADRLDALVAESGDDAVIS----LALDVTDEAQWQEAVERAVT
MUL_1052|M.ulcerans_Agy99 SVAACDLRADRLDALVAESGDDAVIS----LALDVTDEAQWQEAVERAVT
TH_3912|M.thermoresistible__bu RVAACDVLVDELHGSVADLDGDTVLP----LALDVTSAPQWADAVAATVG
:. . . : **:. * .
Mflv_2469|M.gilvum_PYR-GCK ELGPVHILVTSAGLVDFAPFLDISPQLWQRLIDVNLNGTFHCAQAAIPDM
Mvan_4185|M.vanbaalenii_PYR-1 ELGPVHILVTSAGLVDFAPFLEISPDRWQRLIEVNLNGTFHCAQVAIPDM
MLBr_01807|M.leprae_Br4923 HQGPVEVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNM
MAB_0952|M.abscessus_ATCC_1997 AFGALDVVCANAGIFPEARLDTMTPEQLSEVLDVNVKGTVYTVQACLAPL
MSMEG_4004|M.smegmatis_MC2_155 TLGSVSVLVSNAMTVDIASLDETSTASWHRQLDVMLTGTFHGVKTFLPDL
Mb2025|M.bovis_AF2122/97 AFGGLHVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPM
Rv2002|M.tuberculosis_H37Rv AFGGLHVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPM
MAV_2509|M.avium_104 EFGRIDVLVNNAGIINIGTLEDYALSEWQRILDINLTGVFLGIRAVVKPM
MMAR_0402|M.marinum_M RFGALTALINNAGVLHRAALVDETAAGFESAWRVNCLGAFLGMRAALGAL
MUL_1052|M.ulcerans_Agy99 RFGALTALINNAGVLHRAALVDETAAGFESAWRVNCLGAFLGMRAALGAL
TH_3912|M.thermoresistible__bu RFGALSALINNAGVLHRAALPDETAEGFERSWRVNCLGPFLGIRAALDPL
* : : .* : : : * . : :
Mflv_2469|M.gilvum_PYR-GCK LAAGWGRIVMISSSS-AQRGSPGMAHYAASKGALISLTRSLAREYGAAGI
Mvan_4185|M.vanbaalenii_PYR-1 LAAGWGRIVMISSSS-AQRGSPGMAHYAASKGALISFTRSLAREYGPAGI
MLBr_01807|M.leprae_Br4923 IKQRFGRYIFMGSVV-GLSGVPGQANYAASKSGLIGMARSLARELGSRSI
MAB_0952|M.abscessus_ATCC_1997 TASGRGRVILTSSITGPVTGYPGWSHYGASKAAQLGFMRTAAIELAPRGV
MSMEG_4004|M.smegmatis_MC2_155 RAAR-GSIVLVSSVH-AMFGLPGRPAYAAAKAGLTGLGRQLAAEYGP-QL
Mb2025|M.bovis_AF2122/97 KEAGRGSIINISSIE-GLAGTVACHGYTATKFAVRGLTKSTALELGPSGI
Rv2002|M.tuberculosis_H37Rv KEAGRGSIINISSIE-GLAGTVACHGYTATKFAVRGLTKSTALELGPSGI
MAV_2509|M.avium_104 KKAGRGSIINISSIE-GMAGTIACHGYTATKFAVRGLTKSAALELGPSGI
MMAR_0402|M.marinum_M SAAEGPVIVNICSTG-AIRPFPHHCAYGAAKWALRGLTQTAAAELAPAGI
MUL_1052|M.ulcerans_Agy99 SAAEGPVIVNICSTG-AIRPFPHHCAYGAAKWALRGLTQTAAAELAPAGF
TH_3912|M.thermoresistible__bu RHAHNASVVNTCSTG-ALRAFPNHAAYGSSKWALRGLTQIAAAELAAAGI
: * * ::* . .: : * * .. .
Mflv_2469|M.gilvum_PYR-GCK TVNNIPPSGIETPMQLQSQAAGFLPPNEQMAASIPVGHLGTGDDIAAAVG
Mvan_4185|M.vanbaalenii_PYR-1 TVNNVPPSGIETPMQHQSQAAGFLPPNEQMAASIPVGHLGTGDDIAAAVG
MLBr_01807|M.leprae_Br4923 TANVVAPGFIDTEMTRAMDSK----YQEAALQAIPLGRIGAVEEIAGAVS
MAB_0952|M.abscessus_ATCC_1997 TVNAILPGNILTEGLVDMGEE----YISGMARSIPMGMLGSPVDIGHLAA
MSMEG_4004|M.smegmatis_MC2_155 RVNTVLPGPILTASWDGIGEQ----DRRTSAAATALGRLGTPREVASVIS
Mb2025|M.bovis_AF2122/97 RVNSIHPGLVKTPMTDWVPED-------IFQT--ALGRAAEPVEVSNLVV
Rv2002|M.tuberculosis_H37Rv RVNSIHPGLVKTPMTDWVPED-------IFQT--ALGRAAEPVEVSNLVV
MAV_2509|M.avium_104 RVNSIHPGLIKTPMTEWVPED-------IFQS--ALGRAAEPKEVSNLVV
MMAR_0402|M.marinum_M RVNAVFPGPIATSMLDEPTQA-------RLAAVSMFGRLGRPAEVADAVA
MUL_1052|M.ulcerans_Agy99 RVNAVFPGPIATSMLDEPTQA-------RLAAVSMFGRLGRHAEVADAVA
TH_3912|M.thermoresistible__bu RVNAVLPGPVATPMLDDATQR-------RLAATMAAGRLGRPTEVADAVA
.* : *. : * * . ::.
Mflv_2469|M.gilvum_PYR-GCK FLCSEQAGFITGQTLGVNGGSVMA-------------------
Mvan_4185|M.vanbaalenii_PYR-1 FLCSEEAGFITGQTLGVNGGSVMA-------------------
MLBr_01807|M.leprae_Br4923 FLASEDAGYISGAVIPVDGGLGMGH------------------
MAB_0952|M.abscessus_ATCC_1997 FLATDEAGYITGQAIVVDGGQVLPESPDAVNP-----------
MSMEG_4004|M.smegmatis_MC2_155 FLASEASSYITGASIPVDGGWSVTKDSS---------------
Mb2025|M.bovis_AF2122/97 YLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
Rv2002|M.tuberculosis_H37Rv YLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
MAV_2509|M.avium_104 YLASDESSYSTGSEFVVDGGTTAGLGHKDFSNVETDAQPDWVT
MMAR_0402|M.marinum_M FLVSENSSFITGSELVVDGGQCLQVP-----------------
MUL_1052|M.ulcerans_Agy99 FLVSENSSFITGSELVVDGGQCLQVP-----------------
TH_3912|M.thermoresistible__bu FLVSEQASFITGSELVVDGGQSLTIG-----------------
:* :: :.: :* : *:**