For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTELKPDDAVPDSSATVTSPTTGEPVKLFSRAWATGIAVRYSMVIVMLLVIAYFSYRSARFATPDNLVTI LVAAAPFALIALGQTLVILTGGIDLSVGSVIAVSAMAGAATAKANPGQVWLTVLVSMLVGLAVGCVNGFL VSRINVPPFIATLGTLTAGSGLAYVIGGGAPINGLPPEFGSIANTKILGLQIPVLLMIIGIIALAIIMKR TTYGMRVYAVGGNRHAAEIAGINAKNVLFSVYAFSGLLAGLSGVMLASRVISGPPNLGQGYELDAIAAVV IGGASLMGGRGTIWGTALGLLMIQTLNNGLDILVVPAYWQDVIKGVLIVAAVAVDVWSSRRRT*
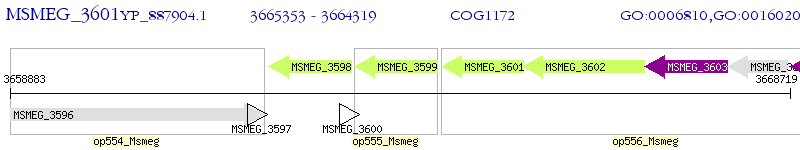
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3601 | - | - | 100% (344) | ribose/xylose/arabinose/galactoside ABC-type transport systems, permease components |
| M. smegmatis MC2 155 | MSMEG_4171 | - | 1e-59 | 38.10% (336) | ribose transport system permease protein RbsC |
| M. smegmatis MC2 155 | MSMEG_6803 | - | 1e-50 | 35.67% (342) | ribose transport system permease protein RbsC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1768c | - | 3e-05 | 22.59% (270) | sulphate-transport transmembrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_2606 | - | 3e-26 | 26.69% (296) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00397 | - | 2e-47 | 35.27% (292) | putative transporter protein |
| M. abscessus ATCC 19977 | MAB_2754 | - | 2e-05 | 26.44% (174) | hypothetical protein MAB_2754 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2329 | - | 1e-15 | 27.11% (284) | branched-chain amino acid ABC-type transport system, permease |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5291 | - | 3e-31 | 26.30% (403) | inner-membrane translocator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3601|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 MDHTTKCDAEQYFQAIVTSMADGVIVVDIDGRIESINPAATRILGLRAHD
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
Mb1768c|M.bovis_AF2122/97 --------------------------------------------------
MAB_2754|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3601|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 VVDMKHGHPFCFYDTDNQRVDLEREVMRVVRREVTTVSKVVGIDQHSGQR
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
Mb1768c|M.bovis_AF2122/97 MIPTMTSAGWAPGVVQFREYQRRWLRGDVLAGLTVAAYLIPQAMAYATVA
MAB_2754|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3601|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 LWLSVNVSLLAYKAPPHSALVVSFSDISAHHLSIERLTYEATHDCLTGLA
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
Mb1768c|M.bovis_AF2122/97 GLPPAAGLWASIAPLAIYALLGSSRQLSIGPESATALMTAAVLAPMAAGD
MAB_2754|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3601|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 NRRFAEDQITKSLQHDERSRLAAVLLLDLDDFKVINDSLGHDVGDAVLQT
Mvan_5291|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
Mb1768c|M.bovis_AF2122/97 LRRYAVLAATLGLLVGLICLLAGTARLGFLASLLSRPVLVGYMAGIALVM
MAB_2754|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3601|M.smegmatis_MC2_155 ----MTTELKPDDAVP----------------------------------
MLBr_00397|M.leprae_Br4923 VAQRLRSAVRPDDVVARLGGDEFIVLLRGPLSDMNANDVAKRLHTTLSES
Mvan_5291|M.vanbaalenii_PYR-1 -----MTTVRSDSDVS----------------------------------
Mflv_2606|M.gilvum_PYR-GCK ------MSTKEALDVG----------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
Mb1768c|M.bovis_AF2122/97 ISSQLGTITGTSVEGNE---------------------------------
MAB_2754|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3601|M.smegmatis_MC2_155 ---------------------DSSATVTSPTTGEPVKLFSRAWATGIAVR
MLBr_00397|M.leprae_Br4923 LVVDQLTVPIGASVGILEVRPDDRRRAADILRDADSAMYAAKNKKQCAVT
Mvan_5291|M.vanbaalenii_PYR-1 ----------------LADADFSGDTRADETFGAAVRGYLQRVRGGDMGS
Mflv_2606|M.gilvum_PYR-GCK --------------------------GHKVVRDERVKERNRLQRLLIRPE
TH_2329|M.thermoresistible__bu -----------------------------------MDILIGQLATGLSLG
Mb1768c|M.bovis_AF2122/97 -------FFSEVHSFATSVTRVHWPTFVLAMSVLALLTMLTRWAPRAPGP
MAB_2754|M.abscessus_ATCC_1997 ---------------------MLYRAFLRVVDLLPMPSLRAQRLIAAAVI
MSMEG_3601|M.smegmatis_MC2_155 YSMVIVMLLVIAYFSYR----SARFATPDNLVTILVAAAPFALIALGQTL
MLBr_00397|M.leprae_Br4923 PQQLVPFVALIALFVFFTAAAGAKFYAPSNLLVILQQTVVLAIVGYGMTF
Mvan_5291|M.vanbaalenii_PYR-1 LPAVLGLVVLFVVFGLAN----DRFLSALNLANLITQAGSICVLAMGLVF
Mflv_2606|M.gilvum_PYR-GCK MGAGIGAIGIFVFFLVVAP----PFREASSLATVLYASSTIGIMACGVAV
TH_2329|M.thermoresistible__bu SILLLAALGLALTFGQMG----------------VINMAHGEFIMVGCYT
Mb1768c|M.bovis_AF2122/97 IIAVLAATMLVAVMSLDAKGIAIVGRIPSGLPTPGVPPVSVEDLRALIIP
MAB_2754|M.abscessus_ATCC_1997 LTQGGIAVTGAVVRVTASGLGCPTWPQCFPGSFTPVAVAEVPRIHQAVEF
:
MSMEG_3601|M.smegmatis_MC2_155 VILTGGIDLSVGSVIAVSAMAGAATAKANPGQVWLTVLVSMLVGLAVGCV
MLBr_00397|M.leprae_Br4923 VIMAGSVDLSVGSIVALTGVTAALVAAQN---QFAAIVTALLVGLAAGMV
Mvan_5291|M.vanbaalenii_PYR-1 VLLLGDIDLSAGVAGGVSACAMALIVVNLSWPWWAALLVGIACGALIGLA
Mflv_2606|M.gilvum_PYR-GCK LMIGGEFDLSAGVAVTFSSLAASMLAYNLHLNLWVGALLALVLALTVGFF
TH_2329|M.thermoresistible__bu AYVVQQIVSDAGVSLLVS--------------LFVGFLVGGAMGALLEVT
Mb1768c|M.bovis_AF2122/97 AAGIAIVTFTDGVLTARAFAARRGQEVNANAELRAVGACNIAAGLTHGFP
MAB_2754|M.abscessus_ATCC_1997 GNRMISFLVVITAALAVLAVTRARRRREVLVYAWLMPASTVLQAIIGGIT
. .
MSMEG_3601|M.smegmatis_MC2_155 NGFLVSRINVPPFIATLGTLTAGSGLAY-VIGGGAPINGLPPEFG-----
MLBr_00397|M.leprae_Br4923 NGIVFAYGKIPSFVSTLGMLQVCRGITL-MISDSSAKP--MPFHG-----
Mvan_5291|M.vanbaalenii_PYR-1 IGLLRAKLGIPSFVVTLAFFLGLQGVTLKLIGEGGSVRVDDPVIRGLTIS
Mflv_2606|M.gilvum_PYR-GCK NGFLVMKTKIPSFLITLSTFFMLAGINLAVTKLVAGQVATQSVSDMQG--
TH_2329|M.thermoresistible__bu LIRRMYHRPLDTLLVTFGVGLILQQVARDIFGAPAVNVLAPAWLGG----
Mb1768c|M.bovis_AF2122/97 VSSSSSRTALADVVGGRTQLYSLIALGLVVIVMVFASGLLAMFPIAALG-
MAB_2754|M.abscessus_ATCC_1997 VRTGLLWWTVAIHLLVSMGMVWLATLLYVKVGEPDVVSDLPSVPP-----
: : :
MSMEG_3601|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 --------------------------------------------------
Mvan_5291|M.vanbaalenii_PYR-1 NLPVAAGWAIAGVVVLGFAGLELHQHRRKIALRLSHSPLGVVVARVGAVA
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
TH_2329|M.thermoresistible__bu --------------------------------------------------
Mb1768c|M.bovis_AF2122/97 --------------------------------------------------
MAB_2754|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3601|M.smegmatis_MC2_155 ---------------SIANTKILGLQIPVLLMIIGIIALAIIMKRTTYGM
MLBr_00397|M.leprae_Br4923 ---------------ILGAMGAMPWILIVCLFVT--ILAGILFQFTMFGR
Mvan_5291|M.vanbaalenii_PYR-1 AVVLGVTYVMSVNRSVSAVADIRGIPYVLPLMLVLLIAMTVVLRRTSYGR
Mflv_2606|M.gilvum_PYR-GCK ------WDSAQKVFASSFTLFGVGIRITVVWWLVFTAVATWVLFKTKVGN
TH_2329|M.thermoresistible__bu ------------GVEIFGAVVPKTRLFILVLAVVCVAALAAVLKYSPAGR
Mb1768c|M.bovis_AF2122/97 ------------ALVVYAALRLIDLSEFRRLARFRRSELMLALATTAAVL
MAB_2754|M.abscessus_ATCC_1997 -----------------------PPAPLSRLTALIGVTLAAVLVAGTLVT
:
MSMEG_3601|M.smegmatis_MC2_155 RVYAVGGNRHAAEIAGINAKN-VLFSVYAFSGLLAGLSGVMLASRVISGP
MLBr_00397|M.leprae_Br4923 WVKAIGGNERVATLAGVPTRG-IKVAIFAICGLTAGLGGIVLASRLGAGT
Mvan_5291|M.vanbaalenii_PYR-1 HIYAVGGNAEAARRAGISVDR-IRVSVFVVCSSLAALSGIIAASYAGKVS
Mflv_2606|M.gilvum_PYR-GCK WIFAVGGDADSARAVGVPVTK-VKIGLFMFVGFCAWFVGMHLLFAFNTVQ
TH_2329|M.thermoresistible__bu RIRAVVQNRDLAETSGISSRT-TDVTTFFIGSGLAAVAGVALT-LIGSTS
Mb1768c|M.bovis_AF2122/97 GLGVFYGVLAAVALSILELLRRVAHPHDSVLGFVPGIAGMHDIDDYPQAK
MAB_2754|M.abscessus_ATCC_1997 GAGPHAGDKSITRVVPR-LQVEITTLVHLHGTLLIAYLALLLGLGFGLAA
. .:
MSMEG_3601|M.smegmatis_MC2_155 PNLGQG-YELDAIAAVVIGGASLMGGRGTIWG------------------
MLBr_00397|M.leprae_Br4923 PTAATG-FEIDVIAAVVIGGTPLTGGLGRLSG------------------
Mvan_5291|M.vanbaalenii_PYR-1 ASSGAGNTLLYAVGAAVIGGTSLFGGKGRAID------------------
Mflv_2606|M.gilvum_PYR-GCK SGQGIG-NEFFYIIAAVIGGCLLTGGYGTAIG------------------
TH_2329|M.thermoresistible__bu PTIGQS-YLIDAFLVVVVG------GLGQIKG------------------
Mb1768c|M.bovis_AF2122/97 RVPGLVVYRYDAPLCFANAEDFRRRALTVVDQDPGQVEWFVLNAESNVEV
MAB_2754|M.abscessus_ATCC_1997 VGVTRHIWVRFTVVLALTLAQALVGVVQFYTG------------------
MSMEG_3601|M.smegmatis_MC2_155 -----TALGLLMIQTLNNGLDILVV--PAYWQDVIKGVLIVAAVAVDVWS
MLBr_00397|M.leprae_Br4923 -----TLIGAIIISMLSNGMVFMGV--GNAASQIIKGIMLAAAVFVFLQR
Mvan_5291|M.vanbaalenii_PYR-1 -----AVIGGVVVATIANGLGLLNQ--SSYINFLVTGGVLLLAASVDAIS
Mflv_2606|M.gilvum_PYR-GCK -----AAIGALIFGMTNQGIVYAGW--DPDWFKFFLGAMLLFAVIANNAF
TH_2329|M.thermoresistible__bu -----TVIAALGLGFLNSFIEYSTT--ASLAKVIVFALIVIFLQVRPQGL
Mb1768c|M.bovis_AF2122/97 DLTALDALDQLRTELLRRGIVFAMARVKQDLRESLRAASLLDKIGEDHIF
MAB_2754|M.abscessus_ATCC_1997 -------VPAVLVALHVAGAAACTAATAALWAALTTPELAATALPKRAEA
: :
MSMEG_3601|M.smegmatis_MC2_155 SRRRT--------
MLBr_00397|M.leprae_Br4923 RKIGIIK------
Mvan_5291|M.vanbaalenii_PYR-1 RRRRSSTGLG---
Mflv_2606|M.gilvum_PYR-GCK RSYAARK------
TH_2329|M.thermoresistible__bu FTVRTRSLA----
Mb1768c|M.bovis_AF2122/97 MTLPTAVQAFRRR
MAB_2754|M.abscessus_ATCC_1997 QPL----------