For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AETRETVVTATAPATPPPPAAKRINLGTLLREPLVGPLVALVIAIIVFSSISNSFLNPQNVSLILQQSVV VGILAVGQTLIILTAGIDLSIGAVAVFGTIVMAQTAGAGGPFLALGATLLVCVAFGAINGGLVTTLRLPP FIVTLGTFTAILAGTRLLAGSETYRVEPGPLTFLGTSFRIGSFSTTYGVVAMLLVYAVVWYALSQTAWGK HVYAVGGNPQSADLLGVKSGRVLFSVYLVTGVIAAIAAWAALGRIPNADPNAYQNANLETITAVVIGGTS LFGGRGGVGGTLVGMLIVAVLRNGLTQAGVDSLYQNIATGILVIVAVAVDQFARRRSR*
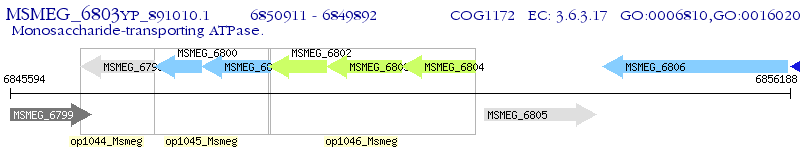
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6803 | - | - | 100% (339) | ribose transport system permease protein RbsC |
| M. smegmatis MC2 155 | MSMEG_3601 | - | 1e-50 | 35.67% (342) | ribose/xylose/arabinose/galactoside ABC-type transport systems, |
| M. smegmatis MC2 155 | MSMEG_4171 | - | 9e-50 | 35.38% (342) | ribose transport system permease protein RbsC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2606 | - | 1e-23 | 26.99% (326) | inner-membrane translocator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00397 | - | 4e-37 | 31.56% (301) | putative transporter protein |
| M. abscessus ATCC 19977 | MAB_2625c | - | 5e-08 | 27.59% (145) | branched chain amino acid ABC transporter |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1081 | - | 3e-08 | 25.95% (158) | ABC transporter branched chain amino acid transport permease |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4010 | - | 2e-23 | 26.38% (326) | inner-membrane translocator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4010|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
MLBr_00397|M.leprae_Br4923 MDHTTKCDAEQYFQAIVTSMADGVIVVDIDGRIESINPAATRILGLRAHD
MSMEG_6803|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4010|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
MLBr_00397|M.leprae_Br4923 VVDMKHGHPFCFYDTDNQRVDLEREVMRVVRREVTTVSKVVGIDQHSGQR
MSMEG_6803|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4010|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2625c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1081|M.thermoresistible__bu --------------------------------------------------
MLBr_00397|M.leprae_Br4923 LWLSVNVSLLAYKAPPHSALVVSFSDISAHHLSIERLTYEATHDCLTGLA
MSMEG_6803|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2606|M.gilvum_PYR-GCK --------------------MSTKEALDVGGHKVVRDER------VKERN
Mvan_4010|M.vanbaalenii_PYR-1 --------------------MSTHADLDVAAHKVVRDER------VKERN
MAB_2625c|M.abscessus_ATCC_199 -------------------MISGWMAQTGVQASTISFDL------HGLWS
TH_1081|M.thermoresistible__bu -------------------MTHQCVALYTCHAADITFNL------QGLRE
MLBr_00397|M.leprae_Br4923 NRRFAEDQITKSLQHDERSRLAAVLLLDLDDFKVINDSLGHDVGDAVLQT
MSMEG_6803|M.smegmatis_MC2_155 -------------------MAETRETVVTATAPATPPPP------AAKRI
Mflv_2606|M.gilvum_PYR-GCK RLQRLLIRPEMGAGIGAIGIFVFFLVVAPPFREASSLATVLYASSTI---
Mvan_4010|M.vanbaalenii_PYR-1 RLQRLLIRPEMGAGIGAIGIFVFFLVVAAPFREASSLATVLYASSTI---
MAB_2625c|M.abscessus_ATCC_199 SIGQLTIDGLAWGAIYALVAVGYTLVFG-------VLRLINFAHSEV---
TH_1081|M.thermoresistible__bu GLLQLTIDGLSWGAIYALVAVGYTLVYG-------VLKLINFAHAEV---
MLBr_00397|M.leprae_Br4923 VAQRLRSAVRPDDVVARLGGDEFIVLLRGPLSDMNANDVAKRLHTTLSES
MSMEG_6803|M.smegmatis_MC2_155 NLGTLLREPLVGPLVALVIAIIVFSSISNSFLNPQNVSLILQQSVVVG--
* : : :
Mflv_2606|M.gilvum_PYR-GCK -----GIMACGVAVLMIGGEFDLSAGVAVTFSSLAASMLAYN--------
Mvan_4010|M.vanbaalenii_PYR-1 -----GIMACGVAVLMIGGEFDLSAGVAVTFSSLAASMLAYN--------
MAB_2625c|M.abscessus_ATCC_199 -----FMLGMFGAYFALDVVLGFSPGGNAYNKGVSLTVLYLG--------
TH_1081|M.thermoresistible__bu -----FMLGMFGSYFCLDVILGFTPRGNAYDLGIGLTVLYLG--------
MLBr_00397|M.leprae_Br4923 LVVDQLTVPIGASVGILEVRPDDRRRAADILRDADSAMYAAKNKKQCAVT
MSMEG_6803|M.smegmatis_MC2_155 ------ILAVGQTLIILTAGIDLSIGAVAVFGTIVMAQTAGAG-------
: : : . :
Mflv_2606|M.gilvum_PYR-GCK ------LHLNLWVGALLALVLALTVGFFNGFLVMKTKIPSFLITLS--TF
Mvan_4010|M.vanbaalenii_PYR-1 ------LHLNLWVGALLALILSLTVGFFNGFLVMRTKIPSFLITLS--TF
MAB_2625c|M.abscessus_ATCC_199 ------VAMLVAMAVSATTALGLEAVAYRPLRRRGARPLTFLITAIGMSF
TH_1081|M.thermoresistible__bu ------IAMLFAIAVSGTAAAGLEFIAYRPLRRRNAARLSFLITAIGMSF
MLBr_00397|M.leprae_Br4923 PQQLVPFVALIALFVFFTAAAGAKFYAPSNLLVILQQTVVLAIVGYGMTF
MSMEG_6803|M.smegmatis_MC2_155 ---------GPFLALGATLLVCVAFGAINGGLVTTLRLPPFIVTLGTFTA
: : : :. :
Mflv_2606|M.gilvum_PYR-GCK FMLAG-------INLAVTKLVAGQVATQ----------------------
Mvan_4010|M.vanbaalenii_PYR-1 FMLAG-------INLAVTKLVAGQVATQ----------------------
MAB_2625c|M.abscessus_ATCC_199 VIQEF-------VHWVLPKIAKGYGGSN----------------------
TH_1081|M.thermoresistible__bu VLQEF-------VHFILPKILDGYGGSN----------------------
MLBr_00397|M.leprae_Br4923 VIMAGSVDLSVGSIVALTGVTAALVAAQNQFAAIVTALLVGLAAGMVNGI
MSMEG_6803|M.smegmatis_MC2_155 ILAG-------------TRLLAGSETYR----------------------
.: . : . .
Mflv_2606|M.gilvum_PYR-GCK -----------SVSDMQGWDSAQKVFASSFTLFG----------VGIRIT
Mvan_4010|M.vanbaalenii_PYR-1 -----------SVSDMQGWDSAQKVFASSFTLFG----------VSIRIT
MAB_2625c|M.abscessus_ATCC_199 -----------AQQPLMLVTPRPVWTFGPFDVSGHHIPFQATITNVTVVV
TH_1081|M.thermoresistible__bu -----------AQQPIRLVTPETQFTLG--DVR---------VSNVTLII
MLBr_00397|M.leprae_Br4923 VFAYGKIPSFVSTLGMLQVCRGITLMISDSSAKPMPFHGILGAMGAMPWI
MSMEG_6803|M.smegmatis_MC2_155 ------------------VEPGPLTFLGTSFRIG---------SFSTTYG
.
Mflv_2606|M.gilvum_PYR-GCK VVWWLVFTAVATWVLFKTKVGNWIFAVGGDADSARAVGVPVTKVKIGLFM
Mvan_4010|M.vanbaalenii_PYR-1 VVWWLVFTAIATWVLFKTKIGNWIFAVGGDADSARAVGVPVTKVKIGLFM
MAB_2625c|M.abscessus_ATCC_199 VAAALVLAFVTDIAINHTKFGRGIRAVAQDSTTATLMGVSRERIIMATFV
TH_1081|M.thermoresistible__bu VFAALVLALITDVVINRTRLGRGIRAVAQDPDTATLMGVSRERVILTTFV
MLBr_00397|M.leprae_Br4923 LIVCLFVTILAGILFQFTMFGRWVKAIGGNERVATLAGVPTRGIKVAIFA
MSMEG_6803|M.smegmatis_MC2_155 VVAMLLVYAVVWYALSQTAWGKHVYAVGGNPQSADLLGVKSGRVLFSVYL
: *.. :. : * *. : *:. : * ** : . :
Mflv_2606|M.gilvum_PYR-GCK FVGFCAWFVGMHLLFAFNTVQSGQGIGNEFFYIIAAVIGGCLLTGGYGTA
Mvan_4010|M.vanbaalenii_PYR-1 FVGFCAWFVGMHLLFAFNTVQSGQGIGNEFFYIIAAVIGGCLLTGGYGTA
MAB_2625c|M.abscessus_ATCC_199 IGGLLAG--AAALLYTLKVPQGIIYSG-GFLLGIKAFS--AAVLGGIGNL
TH_1081|M.thermoresistible__bu IGGLLAG--AAALLYTLKMPQAIIYSG-GFLLGIKAFT--AAVLGGIGNL
MLBr_00397|M.leprae_Br4923 ICGLTAGLGGIVLASRLGAGTPTAATGFEIDVIAAVVIGGTPLTGGLGRL
MSMEG_6803|M.smegmatis_MC2_155 VTGVIAAIAAWAALGRIPNADPNAYQNANLETITAVVIGGTSLFGGRGGV
. *. * . : . : .. : ** *
Mflv_2606|M.gilvum_PYR-GCK IGAAIGALIFGMTN---QGIVYAGWDPDWFKFFLGAMLLFAVIANNAFRS
Mvan_4010|M.vanbaalenii_PYR-1 VGAAIGAFIFGMTN---QGIVYAGWDPDWFKFFLGAMLLFAVIANNAFRS
MAB_2625c|M.abscessus_ATCC_199 RGALLGGLILGVMENYGQAVFGTQWR-DVVAFVLLVLVLMFRPTGILGES
TH_1081|M.thermoresistible__bu RGALLGGLLLGVMENYGQAVFGTQWR-DVVAFVLLVLVLFFRPTGILGES
MLBr_00397|M.leprae_Br4923 SGTLIGAIIISMLS---NGMVFMGVGNAASQIIKGIMLAAAVFVFLQRRK
MSMEG_6803|M.smegmatis_MC2_155 GGTLVGMLIVAVLR---NGLTQAGVDSLYQNIATGILVIVAVAVDQFARR
*: :* ::..: :.: : :: . .
Mflv_2606|M.gilvum_PYR-GCK YAARK-
Mvan_4010|M.vanbaalenii_PYR-1 YAAKK-
MAB_2625c|M.abscessus_ATCC_199 LGRAKA
TH_1081|M.thermoresistible__bu LGKARV
MLBr_00397|M.leprae_Br4923 IGIIK-
MSMEG_6803|M.smegmatis_MC2_155 RSR---
.