For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEQTLPEAEHLSRQLSNRHIQLIAIGGAIGTGLFMGSGKTVSLAGPSVIFVYMIIGFMLFFVMRAMGEL LLSNLEYKSFADFAADLLGPWAGFFTGWTYWFCWIVTGVAEVVAIAGYVAFWWPDLPLWIPALVTVAVLI TLNLATVRAFGETEFWFAMIKIVAILALIVVGLVMVLSHFRAPGGAVASLSNLWDDGGFFPTGPMGFVAG FQIATFAFVGIELVGTTAAEARDPERNLPRAVNSIPVRVMLFYVAALVIIIAVTPWREISPDRSPFVAMF SLAGLGIAASVVNFVVLTSAASSANSGIYSTSRMVYGLSCGGDAPASFGRLTSRRVPANALFLSGVVLLS SVVMVAAGDSIIAAFTLVTTISSLCFMFIWTIILSSYLVYRRRRPQQHSASPFKMPGGVVMCWVVLAFFV FLVWAFTQKSDTLQALLVTPAWFVVLGIAWAFLRRRLGHKAREAAFRAALEAESADSAD
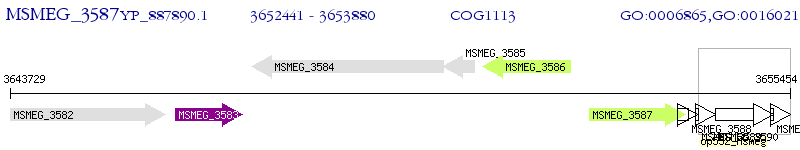
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3587 | - | - | 100% (479) | D-serine/D-alanine/glycine transporter |
| M. smegmatis MC2 155 | MSMEG_5473 | - | 2e-78 | 37.11% (450) | transporter |
| M. smegmatis MC2 155 | MSMEG_6196 | - | 3e-73 | 37.19% (449) | gaba permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1730c | cycA | 1e-173 | 62.53% (451) | D-serine/alanine/glycine transporter protein CycA |
| M. gilvum PYR-GCK | Mflv_1998 | - | 2e-82 | 37.39% (452) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv1704c | cycA | 1e-173 | 62.31% (451) | D-serine/alanine/glycine transporter protein CycA |
| M. leprae Br4923 | MLBr_01305 | ansP2 | 5e-75 | 33.83% (467) | putative L-asparagine permease |
| M. abscessus ATCC 19977 | MAB_3866c | - | 1e-102 | 41.08% (465) | putative amino acid permease |
| M. marinum M | MMAR_1093 | cycA | 1e-179 | 62.53% (467) | D-serine/D-alanine/glycine transporter, CycA |
| M. avium 104 | MAV_4626 | - | 7e-75 | 36.36% (451) | gaba permease |
| M. thermoresistible (build 8) | TH_0471 | - | 4e-18 | 20.92% (459) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_0851 | cycA | 1e-176 | 62.50% (464) | D-serine/D-alanine/glycine transporter, CycA |
| M. vanbaalenii PYR-1 | Mvan_3196 | - | 8e-26 | 25.37% (469) | ethanolamine transproter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1730c|M.bovis_AF2122/97 ---------MPDDIAAADPTDTQPHLRRDLANRHIQLIAIGGA----IGT
Rv1704c|M.tuberculosis_H37Rv ---------MPDDIAAADPTDTQPHLRRDLANRHIQLIAIGGA----IGT
MMAR_1093|M.marinum_M ---------MPEDVATCQPTESTPHLHRGLSNRHIQLIAIGGA----IGT
MUL_0851|M.ulcerans_Agy99 --------MMPEDVATCQHTESTPHLHRGLSNRHIQLIAIGGA----IGT
MSMEG_3587|M.smegmatis_MC2_155 --------------MSEQTLPEAEHLSRQLSNRHIQLIAIGGA----IGT
MAB_3866c|M.abscessus_ATCC_199 -----------MTVANSESETPDAGYSRGLSARTVQMIAIGGA----IGT
MLBr_01305|M.leprae_Br4923 MATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGIGGA----IGT
Mflv_1998|M.gilvum_PYR-GCK -------------------MSAAPTLKKALNQRQLTMIAIGGV----IGA
MAV_4626|M.avium_104 -------------------MAGTSQLRQGLSQRQLSMIALGGV----IGA
Mvan_3196|M.vanbaalenii_PYR-1 -MSGEAHSTHTTRGVEAHRESDAYLQKRQLKKGSAGWILLAGLGISYVVS
TH_0471|M.thermoresistible__bu --MTDRLRVKSVEQSIADTDEPDSRLRKDLSWWDLTVFGVSVV----IGA
: * : :. : :
Mb1730c|M.bovis_AF2122/97 GLFMGSGRTISLAGP-AVMVVYGIIGFFVFFVLRAMGELLLSNLNYKSFV
Rv1704c|M.tuberculosis_H37Rv GLFMGSGRTISLAGP-AVMVVYGIIGFFVFFVLRAMGELLLSNLNYKSFV
MMAR_1093|M.marinum_M GLFMGSGRTISLAGP-AVIVVYAVIGFFVFFVLRAMGELLLSNLNYKSFV
MUL_0851|M.ulcerans_Agy99 GLFMGSGRTISLAGP-AVIVVYAVIGFFVFFVLRAMGELLLSNLNYKSFV
MSMEG_3587|M.smegmatis_MC2_155 GLFMGSGKTVSLAGP-SVIFVYMIIGFMLFFVMRAMGELLLSNLEYKSFA
MAB_3866c|M.abscessus_ATCC_199 GLFYGAGGAIEKAGP-GLILTYLVAGLAIFVIMRALGELLVYRPVSGSIS
MLBr_01305|M.leprae_Br4923 GLFLGAGGRLAKAGP-GLFLVYAVCGVFVFLILRALGELVLHRPSSGSFV
Mflv_1998|M.gilvum_PYR-GCK GLFVGSGVVIADTGP-GAFLTYGMAGVLIIMVMRMLAEMAVANPSTGSFA
MAV_4626|M.avium_104 GLFVGSGVVIKDTGP-AAFLTYALCGLLIVLVMRMLGEMAAANPSTGSFA
Mvan_3196|M.vanbaalenii_PYR-1 GDYSGWNFGLGQGGFGGLAIAAVVIAAMYLAMVLGMAEMSSALPAAGGGY
TH_0471|M.thermoresistible__bu GIFTVTASTAGNITGPAISVSFIIAAIACGLAALCYAEFASTVPVAGSAY
* : . . : . .*: .
Mb1730c|M.bovis_AF2122/97 DFAADLLGPAAGFFVGWSYWFAWVVTGIADLVAITGYARFW--WPGLPIW
Rv1704c|M.tuberculosis_H37Rv DFAADLRGPAAGFFVGWSYWFAWVVTGIADLVAITGYARFW--WPGLPIW
MMAR_1093|M.marinum_M DCAADLLGPAAGYFMGWSYWFAWIVAGIADLVAITGYSKFW--WPDVPTW
MUL_0851|M.ulcerans_Agy99 DCAADLLGPAAGYFMGWSYWFAWIVAGIADLVAITGYSKFW--WPDVPTW
MSMEG_3587|M.smegmatis_MC2_155 DFAADLLGPWAGFFTGWTYWFCWIVTGVAEVVAIAGYVAFW--WPDLPLW
MAB_3866c|M.abscessus_ATCC_199 EYAEEFMGRFAGFANGWTYWAVWATTCMAEITVAGVYIQRW--WPGIPQW
MLBr_01305|M.leprae_Br4923 SYAREFFGEKAAYVVGWLYFLDWAMTAIVDTTAIATYLHRWTIFTALPQW
Mflv_1998|M.gilvum_PYR-GCK DYSRNALGHWAGFSVGWLYWYFWVIVVGFEAIAGAKIVQYW---MDVPLW
MAV_4626|M.avium_104 DYAAAALGRWAGFSVAWLYWYFWVIVVGFEAVAGGKVLNYW---FHAPLW
Mvan_3196|M.vanbaalenii_PYR-1 TFARRALGPWGGFATGTAILIEYSIAPAAIATFIGAYVESLGLFGITDGW
TH_0471|M.thermoresistible__bu TFSYATFGEFVAWIIGWDLILEFSVAAAVVAKGWSSYLGEVFEFAGATTR
: * .: . : .
Mb1730c|M.bovis_AF2122/97 VP-------ALVTVALILAVNLFSVRHFGELEFWFALIKVAAIVCLIAVG
Rv1704c|M.tuberculosis_H37Rv VP-------ALVTVALILAVNLFSVRHFGELEFWFALIKVAAIVCLIAVG
MMAR_1093|M.marinum_M AP-------ALVTIAVMLSINMFGVSKFGETEFWFALIKVAAILCLITVG
MUL_0851|M.ulcerans_Agy99 AP-------ALVTIAVMLSINMFGVSKFGGTEFWFALIKVAAILCLITVG
MSMEG_3587|M.smegmatis_MC2_155 IP-------ALVTVAVLITLNLATVRAFGETEFWFAMIKIVAILALIVVG
MAB_3866c|M.abscessus_ATCC_199 VT-------ALVVLLILFGANLISVKIFGEAEFWFSMIKVTAILGMIIIG
MLBr_01305|M.leprae_Br4923 TL-------ALLALAVVLVMNLISVEWFGELEFWAALIKVCALMAFLVVG
Mflv_1998|M.gilvum_PYR-GCK LS-------ALIFMVLMTATNLFSVASYGEFEFWFAGIKVAAIIAFIAVG
MAV_4626|M.avium_104 VL-------SLGLMLLMTATNLFSVSSFGEFEFWFAGIKVATIVIFLVVG
Mvan_3196|M.vanbaalenii_PYR-1 WV-------YLAAYAIFIGIHLSGVGEALKVMFVITAIALVGLVVFAVAA
TH_0471|M.thermoresistible__bu LGPITLDWGALLIIGFVTVLLATGTKLSANVSLAITVIKVGVVLLVVIVG
* .. . : : * : :: . .
Mb1730c|M.bovis_AF2122/97 A--ILVATNFVSPHGVHATIEN----------LWNDNGFFPTGFLGVVSG
Rv1704c|M.tuberculosis_H37Rv A--ILVATNFVSPHGVHATIEN----------LWNDNGFFPTGFLGVVSG
MMAR_1093|M.marinum_M A--ILVITGFTSPDGDRAALAN----------LWNDGGFFPTGFVGLASG
MUL_0851|M.ulcerans_Agy99 A--ILVITGFTSPDGDRAALAN----------LWNDGGFFPTGFVGLASG
MSMEG_3587|M.smegmatis_MC2_155 L--VMVLSHFRAPGGAVASLSN----------LWDDGGFFPTGPMGFVAG
MAB_3866c|M.abscessus_ATCC_199 VGVLLPISGLGPETG--PSVAN----------LWNDGGVFPTGFSQALLS
MLBr_01305|M.leprae_Br4923 T---IFLGGRYPVDGHNTGLSL----------WTSHGGLFPTGVAPLIVV
Mflv_1998|M.gilvum_PYR-GCK A-----AFVLGLWPGKGADFSN----------LTSHGGFMPLGFGAITVG
MAV_4626|M.avium_104 A-----AYVIGLVPGHHDGLTN----------LVSHGGFFPRGVGAVFAA
Mvan_3196|M.vanbaalenii_PYR-1 IP------HFDSTNLTDIAVTD----------AAGASALLPFGLLGIWAA
TH_0471|M.thermoresistible__bu AFFIKAANYTPFIPPAEAGGEAGRSLDQSLFSLITGAEGSSYGWYGLLAG
. *
Mb1730c|M.bovis_AF2122/97 FQIAFFAYIGVELVGTAAAETADPRRTLPRAINAVPLRVAVFYIGALLAI
Rv1704c|M.tuberculosis_H37Rv FQIAFFAYIGVELVGTAAAETADPRRTLPRAINAVPLRVAVFYIGALLAI
MMAR_1093|M.marinum_M FQIAFFAYIGVELVGAAAAEAADPSHTLPRAINAVPLRVAIFYIGALLAI
MUL_0851|M.ulcerans_Agy99 FQIAFFAYIGVELVGAAAAEAADPSHTLPRAINAVPLRVAIFYIGALLAI
MSMEG_3587|M.smegmatis_MC2_155 FQIATFAFVGIELVGTTAAEARDPERNLPRAVNSIPVRVMLFYVAALVII
MAB_3866c|M.abscessus_ATCC_199 LQIVVFAYVGVELVGVTAGEAENPKVTLRKAINTLPFRIGLFYVGALIVI
MLBr_01305|M.leprae_Br4923 SSGVMFAYAAVELVGTAAGETVEPKKIMPRAINSVIARIAIFYVGSVILL
Mflv_1998|M.gilvum_PYR-GCK IVTVIFSMVGAEIATIAAAESQDPERAVAKAANSVILRILIFYVGAVLLL
MAV_4626|M.avium_104 IVVVIFSMVGAEIVTVAAAESRDPRRAVQKATRSVVTRIGIFFVGSVFLL
Mvan_3196|M.vanbaalenii_PYR-1 IPFAIWFFLAIEGVPLAAEETAEPERNVPKGIIAGMAVLLVTCVTVLVLT
TH_0471|M.thermoresistible__bu ASIVFFAFIGFDIIATTAEETKNPQRDVARGIMATLGIVTVLYVAVSLVL
. : . : :* *: :* : :. : : : : .
Mb1730c|M.bovis_AF2122/97 LAVVPWRQFASG--ESPFVTMFSLAGLAAAASVVNFVVVTAAASSANSGF
Rv1704c|M.tuberculosis_H37Rv LAVVPWRQFASG--ESPFVTMFSLAGLAAAASVVNFVVVTAAASSANSGF
MMAR_1093|M.marinum_M LSVVPWRRFASG--ESPFVTMFALAGLAAAASVVNFVVITSAASSANSGV
MUL_0851|M.ulcerans_Agy99 LSVVPWRRFASG--ESPFVTMFALAGLAAAASVVNFVVITSAASSANSGV
MSMEG_3587|M.smegmatis_MC2_155 IAVTPWREISPD--RSPFVAMFSLAGLGIAASVVNFVVLTSAASSANSGI
MAB_3866c|M.abscessus_ATCC_199 LSIQGWRNYHKG--ESPFVAVFEYLNIPQAANIVNFILLTAALSSCNSGI
MLBr_01305|M.leprae_Br4923 ALLLPYSAFKAS--ESPFVTFFSKVGFYGAGDLMNIVVLTAALSSLNAGL
Mflv_1998|M.gilvum_PYR-GCK VLIVPWNDESVS--ASPFVAAFNAMGIPYADHVMNAVVLTAVLSCLNSGM
MAV_4626|M.avium_104 VAILPWDSVELG--ASPYVAALKHLGLAGADQVMNAVVLTAVLSCLNSGL
Mvan_3196|M.vanbaalenii_PYR-1 TGAGGAAAMSES--GNPLVEALGDS---GWAKVVNYIGLAGLIASFFSII
TH_0471|M.thermoresistible__bu TGMVHYTELRAAGENANLATAFAANGIDWAAKVISIGALAGLTTVVIVLV
. : ::. ::. : .
Mb1730c|M.bovis_AF2122/97 FSTGRMLFGLADEGHAPAAFHQLNRGGVPAPALLLTAPLLLTSIPLLYAG
Rv1704c|M.tuberculosis_H37Rv FSTGRMLFGLADEGHAPAAFHQLNRGGVPAPALLLTAPLLLTSIPLLYAG
MMAR_1093|M.marinum_M FSTGRMLYGLADEGSAPGIFGRLNRRGVPAPALILTAALLLSAIPLLYAG
MUL_0851|M.ulcerans_Agy99 FSTGRMLYGLADEGSAPGIFGRLNSRGVPAPALILTAALLLSAIPLLYAG
MSMEG_3587|M.smegmatis_MC2_155 YSTSRMVYGLSCGGDAPASFGRLTSRRVPANALFLSGVVLLSSVVMVAAG
MAB_3866c|M.abscessus_ATCC_199 YSTGRMVRSLAQRGDAPAGLQALSSRHVPMLAICFSALAMG--IGVFVNW
MLBr_01305|M.leprae_Br4923 YATGRVMHSIAINGSGPKFTARMSKNGVPYGGILLAAVICLCGVALNAFN
Mflv_1998|M.gilvum_PYR-GCK YTASRMLFVLAARREAPAALVKVTNRGVPANAILASSVVGFLCVIAAAVS
MAV_4626|M.avium_104 YTASRMLFVLAERGEAPARLVRLSGRGVPHIAILCSSAVGFLCVVMARVA
Mvan_3196|M.vanbaalenii_PYR-1 YAYSRQLFALSRAGYLPTVLSVTNSRKAPTLALIVPGVIGFLLSLIG---
TH_0471|M.thermoresistible__bu LGQTRVLFAMSRDGLLPRPLSKTGPRGTPVRITVLVGALVAIAASVFPVG
* : :: * .* .
Mb1730c|M.bovis_AF2122/97 RSVIGAFTLVTTVSSLLFMFVWAMIIISYLVYRRRHPQ-RHTDSVYKMPG
Rv1704c|M.tuberculosis_H37Rv RSVIGAFTLVTTVSSLLFMFVWAMIIISYLVYRRRHPQ-RHTDSVYKMPG
MMAR_1093|M.marinum_M GSVIGAFTLITTISSLLFMFVWTMIVISYLVYRRRHAQ-RHADSAYKMPG
MUL_0851|M.ulcerans_Agy99 GSVTGAFTLITTISSLLFMFVWTMIVISYLVYRRRHAQ-RHADSAYKMPG
MSMEG_3587|M.smegmatis_MC2_155 DSIIAAFTLVTTISSLCFMFIWTIILSSYLVYRRRRPQ-QHSASPFKMPG
MAB_3866c|M.abscessus_ATCC_199 LSPDKAFAYITSVSTIGIIFVWGSILVSHMIYRKRVADGLLPASDYKLPG
MLBr_01305|M.leprae_Br4923 PG--QAFEIVLSVAALGIIAGWGTIVLCQLRLHKMAKAGIMRRPRFRMPL
Mflv_1998|M.gilvum_PYR-GCK PD--TIFAFLLNSSGAIILFVYLLIAISQIVLRYRTPDSKLRVKMWLFP-
MAV_4626|M.avium_104 PN--TVFLFLLNSSGAIILFVYWLIALSQIVLRRRTPAERLSVKMWFFP-
Mvan_3196|M.vanbaalenii_PYR-1 -----QGALLLNMAVFGAALSYVLMMVSHIVLRRREPD---MPRPYRTPG
TH_0471|M.thermoresistible__bu N--------LEEMVNIGTLFAFALVSAGVIVLRRTRPD---LKRGFRVPF
: : : : : : *
Mb1730c|M.bovis_AF2122/97 GVVMCWAVLVFFAFVIWTLT----TETETATALAWFPLWFVLLAVGWLVT
Rv1704c|M.tuberculosis_H37Rv GVVMCWAVLVFFAFVIWTLT----TETETATALAWFPLWFVLLAVGWLVT
MMAR_1093|M.marinum_M GVVMCWAILGFFVFVIWTLT----TETETATALVWFPLWFVILGIGWALV
MUL_0851|M.ulcerans_Agy99 GVVMCWAILGFFVFVIWTLTTETETETETATALVWFPLWFVILGIGWALV
MSMEG_3587|M.smegmatis_MC2_155 GVVMCWVVLAFFVFLVWAFT----QKSDTLQALLVTPAWFVVLGIAWAFL
MAB_3866c|M.abscessus_ATCC_199 APVTNILALGFLALVVLLLF----FTEDGRTAILVGVVWFIIVVLGYFVH
MLBr_01305|M.leprae_Br4923 APYSGYLTLAFLFAVLVVMA---FDKPIGTWTVASLIVIVPALIAGWYSI
Mflv_1998|M.gilvum_PYR-GCK -VLSGLTALGILAILVQMFIDTALRSQLVLSLLSWAVVIALFAANKWFVK
MAV_4626|M.avium_104 -VLSVLTLAGITAVLVQMAFDATARSQLWLSLLSWAVVVALY-----FVA
Mvan_3196|M.vanbaalenii_PYR-1 GVVTTGFALVIAVLAVIATF-----LVDSVAALCCLVVFAAFMAYFGIYS
TH_0471|M.thermoresistible__bu VPWLPLAAIAACLWLMLNLT--------GLTWVRFLGWMALGLVLYFFYG
: :
Mb1730c|M.bovis_AF2122/97 QRRQSRRSFGFH-----CQVVGVRQQLGRGMARLAMKIHARPKLRSAVVV
Rv1704c|M.tuberculosis_H37Rv QRRQSRRSFGFH-----CQVVGVRQQLGRGMARLAMKIHARPKLRSAVVV
MMAR_1093|M.marinum_M RRRP-----------------GHLEQYHR----FQAEMNG--------TT
MUL_0851|M.ulcerans_Agy99 RRRP-----------------GHLEQYHR----FQAEMNG--------TA
MSMEG_3587|M.smegmatis_MC2_155 RRRL-----------------GHKAREAA----FRAALEA----------
MAB_3866c|M.abscessus_ATCC_199 HGEP-----------------AHRETQDA---------------------
MLBr_01305|M.leprae_Br4923 RKRV-----------------MTIARERMGYTGPFPAIAN----------
Mflv_1998|M.gilvum_PYR-GCK KRPAEGDSGAPTGPPHRVLVLANRTVDSDELLAELSRIGADQTAEYLVVV
MAV_4626|M.avium_104 R--SRGRVAAR---------------------------------------
Mvan_3196|M.vanbaalenii_PYR-1 RHHLVAN---------------SPDEEFAALAAAEGELK-----------
TH_0471|M.thermoresistible__bu RHHS------------------MLGRGMTDISARRTG-------------
:
Mb1730c|M.bovis_AF2122/97 EPVSAGEPGARRSAKSVRKLASDDSQSAHCPVAVVGLADGGRDPQYHHDG
Rv1704c|M.tuberculosis_H37Rv EPVSAGEPGARRSAKSVRKLASDDSQSAHCPVAVVGLADGGRDPQYHHDG
MMAR_1093|M.marinum_M EGILPPDASART--------------------------------------
MUL_0851|M.ulcerans_Agy99 EGVLPPDASART--------------------------------------
MSMEG_3587|M.smegmatis_MC2_155 ESADSAD-------------------------------------------
MAB_3866c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01305|M.leprae_Br4923 PPVQPSERSHSQNP------------------------------------
Mflv_1998|M.gilvum_PYR-GCK PASAVDTGAAATHGPRDVTEATQEAATARLESTLAALRGKNLTVQGELGD
MAV_4626|M.avium_104 --------------------------------------------------
Mvan_3196|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0471|M.thermoresistible__bu --------------------------------------------------
Mb1730c|M.bovis_AF2122/97 PDR-----------------------------------------------
Rv1704c|M.tuberculosis_H37Rv PDR-----------------------------------------------
MMAR_1093|M.marinum_M --------------------------------------------------
MUL_0851|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3587|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3866c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01305|M.leprae_Br4923 --------------------------------------------------
Mflv_1998|M.gilvum_PYR-GCK YRPLRALDHAVAEFGPDQIVIATLPPEFSMWHRFDVVDRARAQFTVPVTH
MAV_4626|M.avium_104 --------------------------------------------------
Mvan_3196|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0471|M.thermoresistible__bu --------------------------------------------------
Mb1730c|M.bovis_AF2122/97 -------------
Rv1704c|M.tuberculosis_H37Rv -------------
MMAR_1093|M.marinum_M -------------
MUL_0851|M.ulcerans_Agy99 -------------
MSMEG_3587|M.smegmatis_MC2_155 -------------
MAB_3866c|M.abscessus_ATCC_199 -------------
MLBr_01305|M.leprae_Br4923 -------------
Mflv_1998|M.gilvum_PYR-GCK VVATSAVGNQVPR
MAV_4626|M.avium_104 -------------
Mvan_3196|M.vanbaalenii_PYR-1 -------------
TH_0471|M.thermoresistible__bu -------------