For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDDIAAADPTDTQPHLRRDLANRHIQLIAIGGAIGTGLFMGSGRTISLAGPAVMVVYGIIGFFVFFVLRA MGELLLSNLNYKSFVDFAADLRGPAAGFFVGWSYWFAWVVTGIADLVAITGYARFWWPGLPIWVPALVTV ALILAVNLFSVRHFGELEFWFALIKVAAIVCLIAVGAILVATNFVSPHGVHATIENLWNDNGFFPTGFLG VVSGFQIAFFAYIGVELVGTAAAETADPRRTLPRAINAVPLRVAVFYIGALLAILAVVPWRQFASGESPF VTMFSLAGLAAAASVVNFVVVTAAASSANSGFFSTGRMLFGLADEGHAPAAFHQLNRGGVPAPALLLTAP LLLTSIPLLYAGRSVIGAFTLVTTVSSLLFMFVWAMIIISYLVYRRRHPQRHTDSVYKMPGGVVMCWAVL VFFAFVIWTLTTETETATALAWFPLWFVLLAVGWLVTQRRQSRRSFGFHCQVVGVRQQLGRGMARLAMKI HARPKLRSAVVVEPVSAGEPGARRSAKSVRKLASDDSQSAHCPVAVVGLADGGRDPQYHHDGPDR*
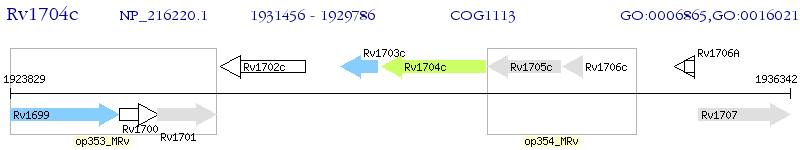
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1704c | cycA | - | 100% (556) | PROBABLE D-SERINE/ALANINE/GLYCINE TRANSPORTER PROTEIN CYCA |
| M. tuberculosis H37Rv | Rv2127 | ansP1 | 7e-86 | 38.63% (453) | L-asparagine permease ansP1 |
| M. tuberculosis H37Rv | Rv0346c | ansP2 | 1e-81 | 39.91% (431) | L-asparagine ABC transporter permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1730c | cycA | 0.0 | 99.82% (556) | D-serine/alanine/glycine transporter protein CycA |
| M. gilvum PYR-GCK | Mflv_1998 | - | 6e-77 | 36.30% (449) | amino acid permease-associated region |
| M. leprae Br4923 | MLBr_01305 | ansP2 | 4e-81 | 37.47% (451) | putative L-asparagine permease |
| M. abscessus ATCC 19977 | MAB_3866c | - | 4e-99 | 41.43% (449) | putative amino acid permease |
| M. marinum M | MMAR_1093 | cycA | 0.0 | 80.04% (461) | D-serine/D-alanine/glycine transporter, CycA |
| M. avium 104 | MAV_4626 | - | 4e-75 | 37.03% (451) | gaba permease |
| M. smegmatis MC2 155 | MSMEG_3587 | - | 1e-173 | 62.31% (451) | D-serine/D-alanine/glycine transporter |
| M. thermoresistible (build 8) | TH_0471 | - | 1e-19 | 22.43% (477) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_0851 | cycA | 0.0 | 78.49% (465) | D-serine/D-alanine/glycine transporter, CycA |
| M. vanbaalenii PYR-1 | Mvan_1740 | - | 2e-23 | 21.34% (492) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1704c|M.tuberculosis_H37Rv ---------------MPDDIAAADPTDTQPHLRRDLANRHIQLIAIGGAI
Mb1730c|M.bovis_AF2122/97 ---------------MPDDIAAADPTDTQPHLRRDLANRHIQLIAIGGAI
MMAR_1093|M.marinum_M ---------------MPEDVATCQPTESTPHLHRGLSNRHIQLIAIGGAI
MUL_0851|M.ulcerans_Agy99 --------------MMPEDVATCQHTESTPHLHRGLSNRHIQLIAIGGAI
MSMEG_3587|M.smegmatis_MC2_155 --------------------MSEQTLPEAEHLSRQLSNRHIQLIAIGGAI
MAB_3866c|M.abscessus_ATCC_199 -----------------MTVANSESETPDAGYSRGLSARTVQMIAIGGAI
MLBr_01305|M.leprae_Br4923 ------MATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGIGGAI
Mflv_1998|M.gilvum_PYR-GCK -------------------------MSAAPTLKKALNQRQLTMIAIGGVI
MAV_4626|M.avium_104 -------------------------MAGTSQLRQGLSQRQLSMIALGGVI
TH_0471|M.thermoresistible__bu --------MTDRLRVKSVEQSIADTDEPDSRLRKDLSWWDLTVFGVSVVI
Mvan_1740|M.vanbaalenii_PYR-1 MVQEPGAQTVGRRRTKSVEQSIADTDEPSTRLRKELNWWDLTVFGVSVVI
: * : ::.:. .*
Rv1704c|M.tuberculosis_H37Rv GTGLFMGSGRTIS-LAGPAVMVVYGIIGFFVFFVLRAMGELLLSNLNYKS
Mb1730c|M.bovis_AF2122/97 GTGLFMGSGRTIS-LAGPAVMVVYGIIGFFVFFVLRAMGELLLSNLNYKS
MMAR_1093|M.marinum_M GTGLFMGSGRTIS-LAGPAVIVVYAVIGFFVFFVLRAMGELLLSNLNYKS
MUL_0851|M.ulcerans_Agy99 GTGLFMGSGRTIS-LAGPAVIVVYAVIGFFVFFVLRAMGELLLSNLNYKS
MSMEG_3587|M.smegmatis_MC2_155 GTGLFMGSGKTVS-LAGPSVIFVYMIIGFMLFFVMRAMGELLLSNLEYKS
MAB_3866c|M.abscessus_ATCC_199 GTGLFYGAGGAIE-KAGPGLILTYLVAGLAIFVIMRALGELLVYRPVSGS
MLBr_01305|M.leprae_Br4923 GTGLFLGAGGRLA-KAGPGLFLVYAVCGVFVFLILRALGELVLHRPSSGS
Mflv_1998|M.gilvum_PYR-GCK GAGLFVGSGVVIA-DTGPGAFLTYGMAGVLIIMVMRMLAEMAVANPSTGS
MAV_4626|M.avium_104 GAGLFVGSGVVIK-DTGPAAFLTYALCGLLIVLVMRMLGEMAAANPSTGS
TH_0471|M.thermoresistible__bu GAGIFTVTASTAGNITGPAISVSFIIAAIACGLAALCYAEFASTVPVAGS
Mvan_1740|M.vanbaalenii_PYR-1 GAGIFTITASTAANITGPAISLSFIIAAIACGLAALCYAEFASTVPVAGS
*:*:* :. :**. . : : .. . .*: *
Rv1704c|M.tuberculosis_H37Rv FVDFAADLRGPAAGFFVGWSYWFAWVVTGIADLVAITGYARFW--WPGLP
Mb1730c|M.bovis_AF2122/97 FVDFAADLLGPAAGFFVGWSYWFAWVVTGIADLVAITGYARFW--WPGLP
MMAR_1093|M.marinum_M FVDCAADLLGPAAGYFMGWSYWFAWIVAGIADLVAITGYSKFW--WPDVP
MUL_0851|M.ulcerans_Agy99 FVDCAADLLGPAAGYFMGWSYWFAWIVAGIADLVAITGYSKFW--WPDVP
MSMEG_3587|M.smegmatis_MC2_155 FADFAADLLGPWAGFFTGWTYWFCWIVTGVAEVVAIAGYVAFW--WPDLP
MAB_3866c|M.abscessus_ATCC_199 ISEYAEEFMGRFAGFANGWTYWAVWATTCMAEITVAGVYIQRW--WPGIP
MLBr_01305|M.leprae_Br4923 FVSYAREFFGEKAAYVVGWLYFLDWAMTAIVDTTAIATYLHRWTIFTALP
Mflv_1998|M.gilvum_PYR-GCK FADYSRNALGHWAGFSVGWLYWYFWVIVVGFEAIAGAKIVQYW---MDVP
MAV_4626|M.avium_104 FADYAAAALGRWAGFSVAWLYWYFWVIVVGFEAVAGGKVLNYW---FHAP
TH_0471|M.thermoresistible__bu AYTFSYATFGEFVAWIIGWDLILEFSVAAAVVAKGWSSYLGEVFEFAGAT
Mvan_1740|M.vanbaalenii_PYR-1 AYTFSYATFGEFVAWIIGWDLILEFAVAAAVVAKGWSSYLGTVFGFAGGT
: * ..: .* : . .
Rv1704c|M.tuberculosis_H37Rv IWVP-------ALVTVALILAVNLFSVRHFGELEFWFALIKVAAIVCLIA
Mb1730c|M.bovis_AF2122/97 IWVP-------ALVTVALILAVNLFSVRHFGELEFWFALIKVAAIVCLIA
MMAR_1093|M.marinum_M TWAP-------ALVTIAVMLSINMFGVSKFGETEFWFALIKVAAILCLIT
MUL_0851|M.ulcerans_Agy99 TWAP-------ALVTIAVMLSINMFGVSKFGGTEFWFALIKVAAILCLIT
MSMEG_3587|M.smegmatis_MC2_155 LWIP-------ALVTVAVLITLNLATVRAFGETEFWFAMIKIVAILALIV
MAB_3866c|M.abscessus_ATCC_199 QWVT-------ALVVLLILFGANLISVKIFGEAEFWFSMIKVTAILGMII
MLBr_01305|M.leprae_Br4923 QWTL-------ALLALAVVLVMNLISVEWFGELEFWAALIKVCALMAFLV
Mflv_1998|M.gilvum_PYR-GCK LWLS-------ALIFMVLMTATNLFSVASYGEFEFWFAGIKVAAIIAFIA
MAV_4626|M.avium_104 LWVL-------SLGLMLLMTATNLFSVSSFGEFEFWFAGIKVATIVIFLV
TH_0471|M.thermoresistible__bu TRLGPITLDWGALLIIGFVTVLLATGTKLSANVSLAITVIKVGVVLLVVI
Mvan_1740|M.vanbaalenii_PYR-1 TEVAGFQLDWGALVIIALVTLILVMGTKLSAGVSLAITAVKVSVVLLVVI
:* : .: . . .: : :*: .:: .:
Rv1704c|M.tuberculosis_H37Rv VGA------------ILVATNFVSPHGVHATIENLWNDNGFFPTGFLGVV
Mb1730c|M.bovis_AF2122/97 VGA------------ILVATNFVSPHGVHATIENLWNDNGFFPTGFLGVV
MMAR_1093|M.marinum_M VGA------------ILVITGFTSPDGDRAALANLWNDGGFFPTGFVGLA
MUL_0851|M.ulcerans_Agy99 VGA------------ILVITGFTSPDGDRAALANLWNDGGFFPTGFVGLA
MSMEG_3587|M.smegmatis_MC2_155 VGL------------VMVLSHFRAPGGAVASLSNLWDDGGFFPTGPMGFV
MAB_3866c|M.abscessus_ATCC_199 IGVG----------VLLPISGLGPETG--PSVANLWNDGGVFPTGFSQAL
MLBr_01305|M.leprae_Br4923 VGT-------------IFLGGRYPVDGHNTGLSLWTSHGGLFPTGVAPLI
Mflv_1998|M.gilvum_PYR-GCK VGA---------------AFVLGLWPGKGADFSNLTSHGGFMPLGFGAIT
MAV_4626|M.avium_104 VGA---------------AYVIGLVPGHHDGLTNLVSHGGFFPRGVGAVF
TH_0471|M.thermoresistible__bu VGAFFIKAANYTPFIPPAEAGGEAGRSLDQSLFSLITGAEGSSYGWYGLL
Mvan_1740|M.vanbaalenii_PYR-1 VGAFYIKAENFSPFVPAPEEGGSGGTGTEQSLFSLLTGAEGSHYGWYGVL
:* . . *
Rv1704c|M.tuberculosis_H37Rv SGFQIAFFAYIGVELVGTAAAETADPRRTLPRAINAVPLRVAVFYIGALL
Mb1730c|M.bovis_AF2122/97 SGFQIAFFAYIGVELVGTAAAETADPRRTLPRAINAVPLRVAVFYIGALL
MMAR_1093|M.marinum_M SGFQIAFFAYIGVELVGAAAAEAADPSHTLPRAINAVPLRVAIFYIGALL
MUL_0851|M.ulcerans_Agy99 SGFQIAFFAYIGVELVGAAAAEAADPSHTLPRAINAVPLRVAIFYIGALL
MSMEG_3587|M.smegmatis_MC2_155 AGFQIATFAFVGIELVGTTAAEARDPERNLPRAVNSIPVRVMLFYVAALV
MAB_3866c|M.abscessus_ATCC_199 LSLQIVVFAYVGVELVGVTAGEAENPKVTLRKAINTLPFRIGLFYVGALI
MLBr_01305|M.leprae_Br4923 VVSSGVMFAYAAVELVGTAAGETVEPKKIMPRAINSVIARIAIFYVGSVI
Mflv_1998|M.gilvum_PYR-GCK VGIVTVIFSMVGAEIATIAAAESQDPERAVAKAANSVILRILIFYVGAVL
MAV_4626|M.avium_104 AAIVVVIFSMVGAEIVTVAAAESRDPRRAVQKATRSVVTRIGIFFVGSVF
TH_0471|M.thermoresistible__bu AGASIVFFAFIGFDIIATTAEETKNPQRDVARGIMATLGIVTVLYVAVSL
Mvan_1740|M.vanbaalenii_PYR-1 AGASIVFFAFIGFDIVATTAEETRNPQRDVPRGILASLAIVTVLYVAVAV
. *: . :: :* *: :* : :. : : ::::. .
Rv1704c|M.tuberculosis_H37Rv AILAVVPWRQFASG--ESPFVTMFSLAGLAAAASVVNFVVVTAAASSANS
Mb1730c|M.bovis_AF2122/97 AILAVVPWRQFASG--ESPFVTMFSLAGLAAAASVVNFVVVTAAASSANS
MMAR_1093|M.marinum_M AILSVVPWRRFASG--ESPFVTMFALAGLAAAASVVNFVVITSAASSANS
MUL_0851|M.ulcerans_Agy99 AILSVVPWRRFASG--ESPFVTMFALAGLAAAASVVNFVVITSAASSANS
MSMEG_3587|M.smegmatis_MC2_155 IIIAVTPWREISPD--RSPFVAMFSLAGLGIAASVVNFVVLTSAASSANS
MAB_3866c|M.abscessus_ATCC_199 VILSIQGWRNYHKG--ESPFVAVFEYLNIPQAANIVNFILLTAALSSCNS
MLBr_01305|M.leprae_Br4923 LLALLLPYSAFKAS--ESPFVTFFSKVGFYGAGDLMNIVVLTAALSSLNA
Mflv_1998|M.gilvum_PYR-GCK LLVLIVPWNDESVS--ASPFVAAFNAMGIPYADHVMNAVVLTAVLSCLNS
MAV_4626|M.avium_104 LLVAILPWDSVELG--ASPYVAALKHLGLAGADQVMNAVVLTAVLSCLNS
TH_0471|M.thermoresistible__bu VLTGMVHYTELRAAGENANLATAFAANGIDWAAKVISIGALAGLTTVVIV
Mvan_1740|M.vanbaalenii_PYR-1 VLTGMVSYTELRDA-ETQNLATAFSLNGVDWAAKVISIGALAGLTTVVIV
: : : .: : .. * ::. ::. :
Rv1704c|M.tuberculosis_H37Rv GFFSTGRMLFGLADEGHAPAAFHQLNRGGVPAPALLLTAPLLLTSIPLLY
Mb1730c|M.bovis_AF2122/97 GFFSTGRMLFGLADEGHAPAAFHQLNRGGVPAPALLLTAPLLLTSIPLLY
MMAR_1093|M.marinum_M GVFSTGRMLYGLADEGSAPGIFGRLNRRGVPAPALILTAALLLSAIPLLY
MUL_0851|M.ulcerans_Agy99 GVFSTGRMLYGLADEGSAPGIFGRLNSRGVPAPALILTAALLLSAIPLLY
MSMEG_3587|M.smegmatis_MC2_155 GIYSTSRMVYGLSCGGDAPASFGRLTSRRVPANALFLSGVVLLSSVVMVA
MAB_3866c|M.abscessus_ATCC_199 GIYSTGRMVRSLAQRGDAPAGLQALSSRHVPMLAICFSALAMG--IGVFV
MLBr_01305|M.leprae_Br4923 GLYATGRVMHSIAINGSGPKFTARMSKNGVPYGGILLAAVICLCGVALNA
Mflv_1998|M.gilvum_PYR-GCK GMYTASRMLFVLAARREAPAALVKVTNRGVPANAILASSVVGFLCVIAAA
MAV_4626|M.avium_104 GLYTASRMLFVLAERGEAPARLVRLSGRGVPHIAILCSSAVGFLCVVMAR
TH_0471|M.thermoresistible__bu LVLGQTRVLFAMSRDGLLPRPLSKTGPRGTPVRITVLVGALVAIAASVFP
Mvan_1740|M.vanbaalenii_PYR-1 LVLGQTRVLFAMSRDGLLPRQLAKTGEHGTPVRITLIVGAVVAVTATVFP
. *:: :: * .* .
Rv1704c|M.tuberculosis_H37Rv AGRSVIGAFTLVTTVSSLLFMFVWAMIIISYLVYRRRHPQ-RHTDSVYKM
Mb1730c|M.bovis_AF2122/97 AGRSVIGAFTLVTTVSSLLFMFVWAMIIISYLVYRRRHPQ-RHTDSVYKM
MMAR_1093|M.marinum_M AGGSVIGAFTLITTISSLLFMFVWTMIVISYLVYRRRHAQ-RHADSAYKM
MUL_0851|M.ulcerans_Agy99 AGGSVTGAFTLITTISSLLFMFVWTMIVISYLVYRRRHAQ-RHADSAYKM
MSMEG_3587|M.smegmatis_MC2_155 AGDSIIAAFTLVTTISSLCFMFIWTIILSSYLVYRRRRPQ-QHSASPFKM
MAB_3866c|M.abscessus_ATCC_199 NWLSPDKAFAYITSVSTIGIIFVWGSILVSHMIYRKRVADGLLPASDYKL
MLBr_01305|M.leprae_Br4923 FNPG--QAFEIVLSVAALGIIAGWGTIVLCQLRLHKMAKAGIMRRPRFRM
Mflv_1998|M.gilvum_PYR-GCK VSPD--TIFAFLLNSSGAIILFVYLLIAISQIVLRYRTPDSKLRVKMWLF
MAV_4626|M.avium_104 VAPN--TVFLFLLNSSGAIILFVYWLIALSQIVLRRRTPAERLSVKMWFF
TH_0471|M.thermoresistible__bu --------VGNLEEMVNIGTLFAFALVSAGVIVLRRTRPDLKRGFRVPFV
Mvan_1740|M.vanbaalenii_PYR-1 --------IGKLEEMVNIGTLFAFVLVSAGVLVLRRTRPDLKRGFRTPWV
. : : : : : : .
Rv1704c|M.tuberculosis_H37Rv PGGVVMCWAVLVFFAFVIWTLT----TETETATALAWFPLWFVLLAVGWL
Mb1730c|M.bovis_AF2122/97 PGGVVMCWAVLVFFAFVIWTLT----TETETATALAWFPLWFVLLAVGWL
MMAR_1093|M.marinum_M PGGVVMCWAILGFFVFVIWTLT----TETETATALVWFPLWFVILGIGWA
MUL_0851|M.ulcerans_Agy99 PGGVVMCWAILGFFVFVIWTLTTETETETETATALVWFPLWFVILGIGWA
MSMEG_3587|M.smegmatis_MC2_155 PGGVVMCWVVLAFFVFLVWAFT----QKSDTLQALLVTPAWFVVLGIAWA
MAB_3866c|M.abscessus_ATCC_199 PGAPVTNILALGFLALVVLLLF----FTEDGRTAILVGVVWFIIVVLGYF
MLBr_01305|M.leprae_Br4923 PLAPYSGYLTLAFLFAVLVVMA---FDKPIGTWTVASLIVIVPALIAGWY
Mflv_1998|M.gilvum_PYR-GCK P--VLSGLTALGILAILVQMFIDTALRSQLVLSLLSWAVVIALFAANKWF
MAV_4626|M.avium_104 P--VLSVLTLAGITAVLVQMAFDATARSQLWLSLLSWAVVVALY-----F
TH_0471|M.thermoresistible__bu PWLPLAAIAACLWLMLNLTGLT-----WVRFLGWMALGLVLYFFYGRHHS
Mvan_1740|M.vanbaalenii_PYR-1 PLLPILAIAACVWLMLNLTGLT-----WIRFLIWMAIGVVVYFLYGRRHS
* : :
Rv1704c|M.tuberculosis_H37Rv VTQRRQSRRSFGFH----CQVVGVRQQLGRGMARLAMKIHARPKLRSAVV
Mb1730c|M.bovis_AF2122/97 VTQRRQSRRSFGFH----CQVVGVRQQLGRGMARLAMKIHARPKLRSAVV
MMAR_1093|M.marinum_M LVRRRP----------------GHLEQYHR----FQAEMNG--------T
MUL_0851|M.ulcerans_Agy99 LVRRRP----------------GHLEQYHR----FQAEMNG--------T
MSMEG_3587|M.smegmatis_MC2_155 FLRRRL----------------GHKAR--------EAAFRA--------A
MAB_3866c|M.abscessus_ATCC_199 VH-------------------------------------HG---------
MLBr_01305|M.leprae_Br4923 SIRKRVMT--------------------------IARERMGYTGPFPAIA
Mflv_1998|M.gilvum_PYR-GCK VKKRPAEGDSGAPTGPPHRVLVLANRTVDSDELLAELSRIGADQTAEYLV
MAV_4626|M.avium_104 VAR--SRGRVAAR-------------------------------------
TH_0471|M.thermoresistible__bu MLGRGMTD------------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 LVGRRMG-------------------------------------------
Rv1704c|M.tuberculosis_H37Rv VEPVSAGEPGARRSAKSVRKLASDDSQSAHCPVAVVGLADGGRDPQYHHD
Mb1730c|M.bovis_AF2122/97 VEPVSAGEPGARRSAKSVRKLASDDSQSAHCPVAVVGLADGGRDPQYHHD
MMAR_1093|M.marinum_M TEGILPPDASART-------------------------------------
MUL_0851|M.ulcerans_Agy99 AEGVLPPDASART-------------------------------------
MSMEG_3587|M.smegmatis_MC2_155 LEAESADSAD----------------------------------------
MAB_3866c|M.abscessus_ATCC_199 -EPAHRETQDA---------------------------------------
MLBr_01305|M.leprae_Br4923 NPPVQPSERSHSQNP-----------------------------------
Mflv_1998|M.gilvum_PYR-GCK VVPASAVDTGAAATHGPRDVTEATQEAATARLESTLAALRGKNLTVQGEL
MAV_4626|M.avium_104 --------------------------------------------------
TH_0471|M.thermoresistible__bu ---ISARRTG----------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1704c|M.tuberculosis_H37Rv GPDR----------------------------------------------
Mb1730c|M.bovis_AF2122/97 GPDR----------------------------------------------
MMAR_1093|M.marinum_M --------------------------------------------------
MUL_0851|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3587|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3866c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01305|M.leprae_Br4923 --------------------------------------------------
Mflv_1998|M.gilvum_PYR-GCK GDYRPLRALDHAVAEFGPDQIVIATLPPEFSMWHRFDVVDRARAQFTVPV
MAV_4626|M.avium_104 --------------------------------------------------
TH_0471|M.thermoresistible__bu --------------------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1704c|M.tuberculosis_H37Rv ---------------
Mb1730c|M.bovis_AF2122/97 ---------------
MMAR_1093|M.marinum_M ---------------
MUL_0851|M.ulcerans_Agy99 ---------------
MSMEG_3587|M.smegmatis_MC2_155 ---------------
MAB_3866c|M.abscessus_ATCC_199 ---------------
MLBr_01305|M.leprae_Br4923 ---------------
Mflv_1998|M.gilvum_PYR-GCK THVVATSAVGNQVPR
MAV_4626|M.avium_104 ---------------
TH_0471|M.thermoresistible__bu ---------------
Mvan_1740|M.vanbaalenii_PYR-1 ---------------